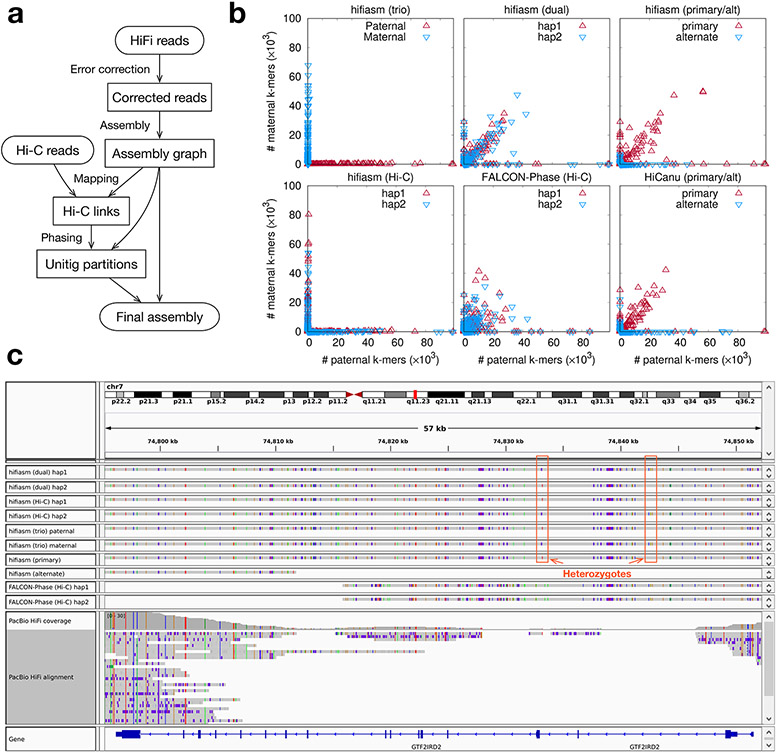
Figure 1. Haplotype-resolved assembly using Hi-C data.
(a) Assembly workflow. Hifiasm corrects reads and produces a phased assembly graph. It then maps Hi-C short reads to the graph, links unitigs in the assembly graph that share mapped Hi-C fragments, and finds a bipartition of unitigs such that unitigs linked by many Hi-C fragments tend to be grouped together. Hifiasm finally emits a haplotype-resolved assembly jointly considering the unitig partition and the assembly graph. (b) Phasing accuracy of HG002 assemblies. Each point corresponds to a contig. Its coordinate gives the number of paternal- and maternal-specific 31-mers on the contig, with these 31-mers derived from parental short reads. Hifiasm (trio): haplotype-resolved hifiasm assembly with trio binning. Hifiasm (dual): paired hifiasm assembly without Hi-C. Hifiasm (primary/alt): primary and alternate hifiasm assembly without Hi-C. Hifiasm (Hi-C): haplotype-resolved hifiasm assembly with Hi-C. FALCON-Phase (Hi-C): FALCON-Phase assembly with Hi-C based on IPA contigs, acquired from its publication10. HiCanu (primary/alt): primary and alternate HiCanu assembly without Hi-C. All assemblies use the same HiFi and Hi-C datasets. (c) Screenshot of contig and read alignment to GRCh38 around gene GTF2IRD2.