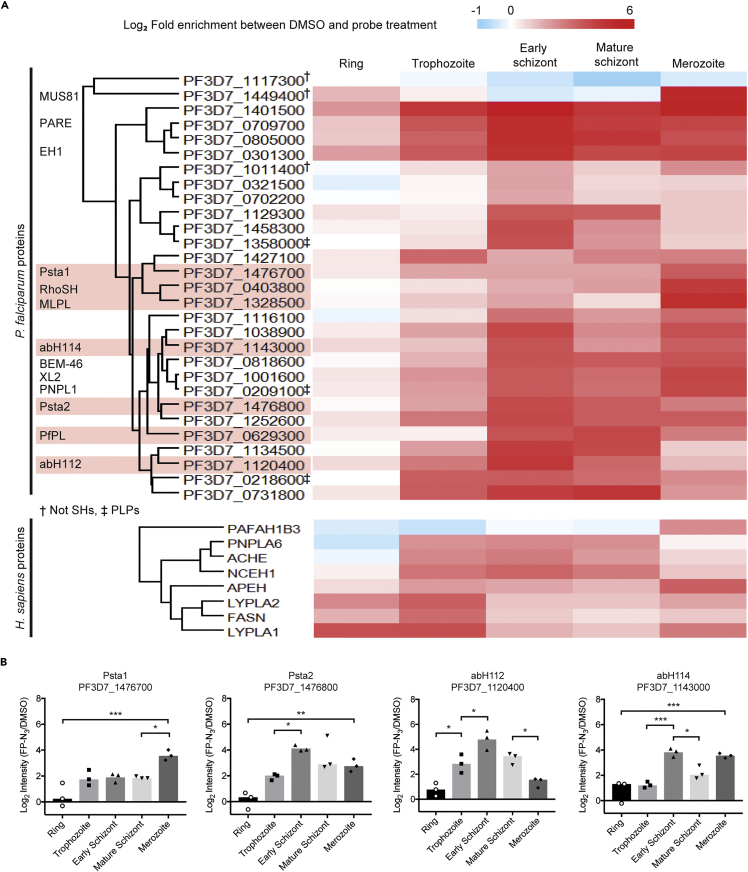
Figure 3.
Activity profile of SHs throughout the erythrocytic cycle
(A) For each protein, fold enrichment was calculated as Log2(Intensity(FP-N3/DMSO)) and used to create a heat map. Proteins were clustered by Euclidean clustering. Proteins discussed in the text are highlighted. † denotes non-SHs, ‡ denotes SHs of PLP-type, all others are α/β-fold hydrolases.
(B) The Log2 enrichment of four selected PfSHs for genetic interrogation are plotted against life cycle stage. The bars represent the mean of three replicates (replicates shown by symbols). Statistical significance was determined by unpaired one-way ANOVA. Significance levels are indicated: p ≤ 0.001, ∗∗∗; p ≤ 0.01, ∗∗; p ≤ 0.05, ∗; p < 0.05, non-significant. See also Table S1.