Fig. 2.
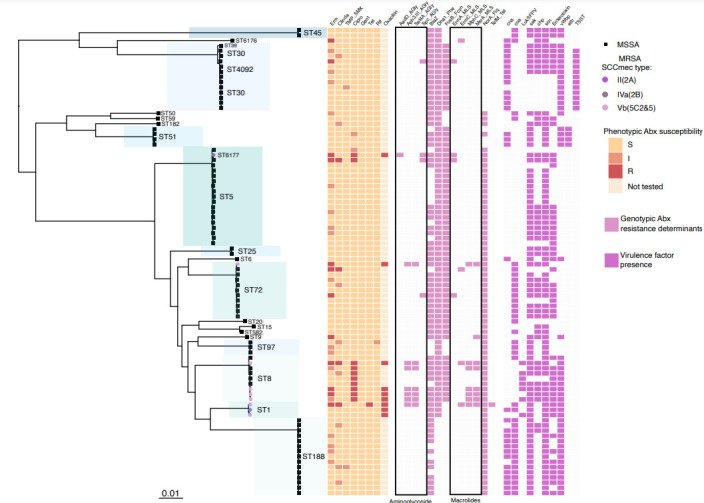
Maximum-likelihood phylogeny illustrating population structure, phenotypic antibiotic susceptibility, and genotypic virulence and antibiotic-resistance determinants. Dominant clades identified through analysis of population structure are highlighted with MLST type annotated. Strains identified as MRSA or MSSA are indicated by circular and square tip shapes, respectively, with SCCmec type shown by the tip colour of MRSA strains. The heatmap to the right shows the phenotypic antibiotic (Abx) susceptibility results, genotypic antibiotic determinants and virulence determinants. Phenotypic antibiotic testing is reported for erythromycin (erm), clindamycin (clinda), trimethoprim/sulfamethoxazole (TMP_SMX), ciprofloxacin (cipro), gentamicin (gent), tetracycline (tet) and oxacillin. The presence of genotypic antibiotic-resistance determinants is indicated by light pink. Antibiotic-resistance determinants include: aminoglycoside resistance (AadD, Aph3.III, Sat4A, Spc), β-lactam resistance (BlaZ, Dha1), fosfomycin resistance (FosfB), macrolide resistance (ErmA, ErmC, MphC, MsrA), fluoroquinolone resistance (NorA) and tetracycline resistance (TetM). Virulence factor presence is indicated in purple, and includes: (i) adherence – collagen binding protein (Cna); (ii) toxin – PVL Panton-Valentine leucocidin (LukF-PV), staphylococcal enterotoxin (enterotoxin), exfoliative toxin b (Etb), toxic shock syndrome toxin-1 (TSST-1); (iii) exoenzyme – von Willebrand factor-binding protein (vWbp), staphylocoagulase (Coa); (iv) immune evasion – CHIPS chemotaxis inhibitory protein of staphylococcus (Chp), SCIN staphylococcal complement inhibitor (Scn); - plasminogen activator – staphylokinase (Sak). The scale bar indicates genetic distance.
