Fig. 1.
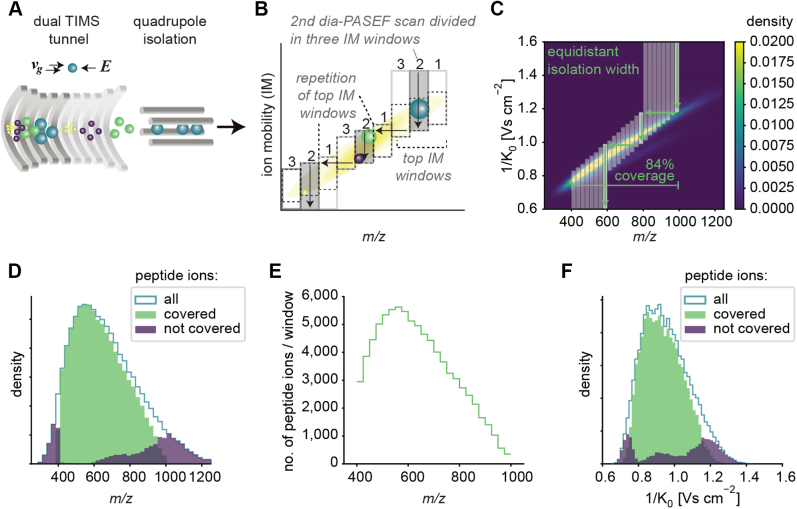
Principle of dia-PASEF on a timsTOF with equidistant two-dimensional isolation windows.A, schematic of a TIMS tunnel followed by quadrupole isolation. B, dia-PASEF acquisition scheme depicting three dia-PASEF scans divided into three ion mobility (IM) windows. Vertical arrows indicate the elution of the ions with decreasing electrical field, and horizontal arrows indicate the movement of the quadrupole. The pattern of the top IM windows is repeated, and the top and bottom IM windows are extended to the upper and lower IM range, respectively. C, original dia-PASEF acquisition scheme (6) plotted on a kernel density distribution of all precursors. One dia-PASEF scan is divided into three IM windows by three distinct movements of quadrupole isolation. This scheme comprises eight dia-PASEF scans with equidistant isolation width covering in total 84% of the peptide ion population. D, histogram of m/z of all peptides covered by the acquisition method in (C), and peptides not covered by the method but identified in a separately recorded spectral library. E, number of peptide ions per isolation window. F, histogram of IMs of all peptides covered by the acquisition method, and peptides not covered by the method but identified in a separately recorded spectral library. The subfigures C–F are based on a reference proteome library (see the Experimental Procedures section). DIA, data-independent acquisition; PASEF, parallel accumulation–serial fragmentation; TIMS, trapped ion mobility spectrometry.