Fig. 2.
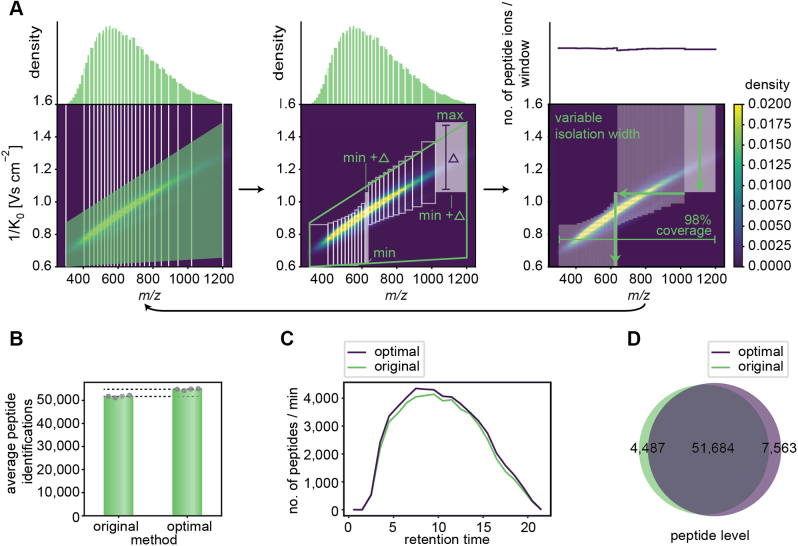
py_diAID algorithm and evaluation.A, py_diAID design of the optimal acquisition scheme and window placement for a 21 min gradient (60 SPD, Evosep) with variable widths to balance the distribution of peptide ions, providing nearly complete peptide ion coverage. The left panel illustrates the first steps of the py_diAID algorithm: defining the m/z range of interest, binning the peptide ions in the m/z dimension and definition of the scan area in the IM dimension. Middle panel, calculation of the isolation window dimensions and coordinates based on the scan area. Right panel, extension of the isolation windows to the limits of the IM ranges. The arrow at the bottom indicates that the py_diAID algorithm evaluates the new acquisition scheme, defines the following test set of scan area parameters by Bayesian optimization, and resumes with the steps in the left panel. This is repeated for a user-defined number of iterations (more details in supplemental Fig. S6). A is plotted on top of a kernel density distribution based on the reference proteome library. B, average peptide identifications by the original and optimal dia-PASEF methods. C, number of peptides identified per minute over the entire retention time. D, Venn diagram showing the shared and unique peptides identified by both methods. Data in B–D are from quadruplicate injections of 200 ng tryptic HeLa digest with a 21 min gradient and analyzed with the reference proteome library. DIA, data-independent acquisition; IM, ion mobility; PASEF, parallel accumulation–serial fragmentation; py_diAID, Python package for DIA with an automated isolation design; SPD, samples per day.