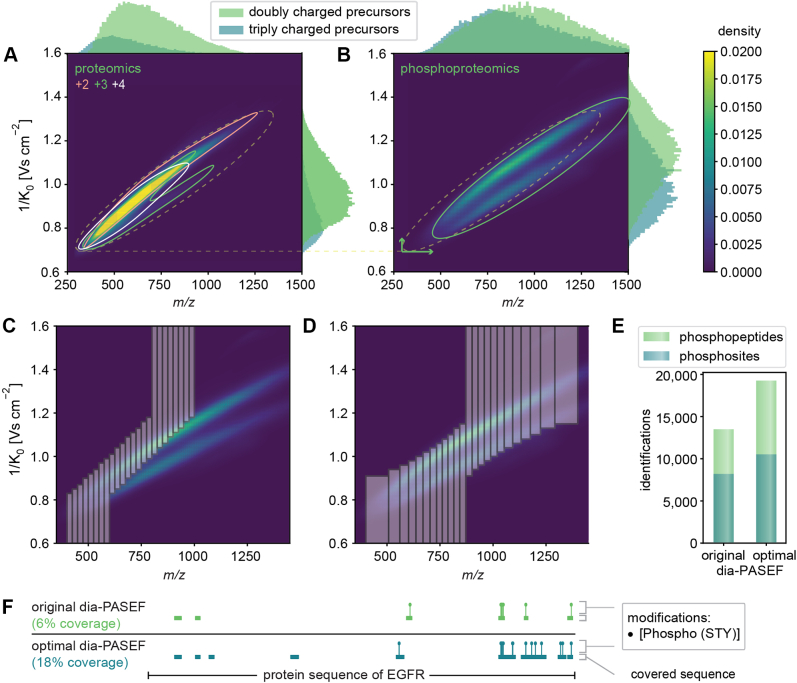
Fig. 5.
Method optimization specifically for phosphoproteomics.A, peptide distribution of a proteomics digest displayed as kernel density estimation dependent on the charge and histograms of the abundance of differently charged precursors based on our deep proteomics library. B, peptide distribution of a phosphoproteomics digest displayed as kernel density estimation and histograms of the abundance of differently charged precursors based on our phosphopeptide library. C, original dia-PASEF method plotted on top of the phosphopeptide library. D, optimal dia-PASEF method tailored to the phospholibrary. E, identified phosphosites and phosphopeptides based on quadruplicates of 100 μg EGF-stimulated and enriched HeLa digest, separated within 21 min, and searched with DIA-NN against the phospholibrary. F, AlphaMap visualization (47): Protein sequence coverage of the EGF receptor (EGFR) depending on the acquisition method. DIA, data-independent acquisition; EGF, epidermal growth factor; PASEF, parallel accumulation–serial fragmentation.