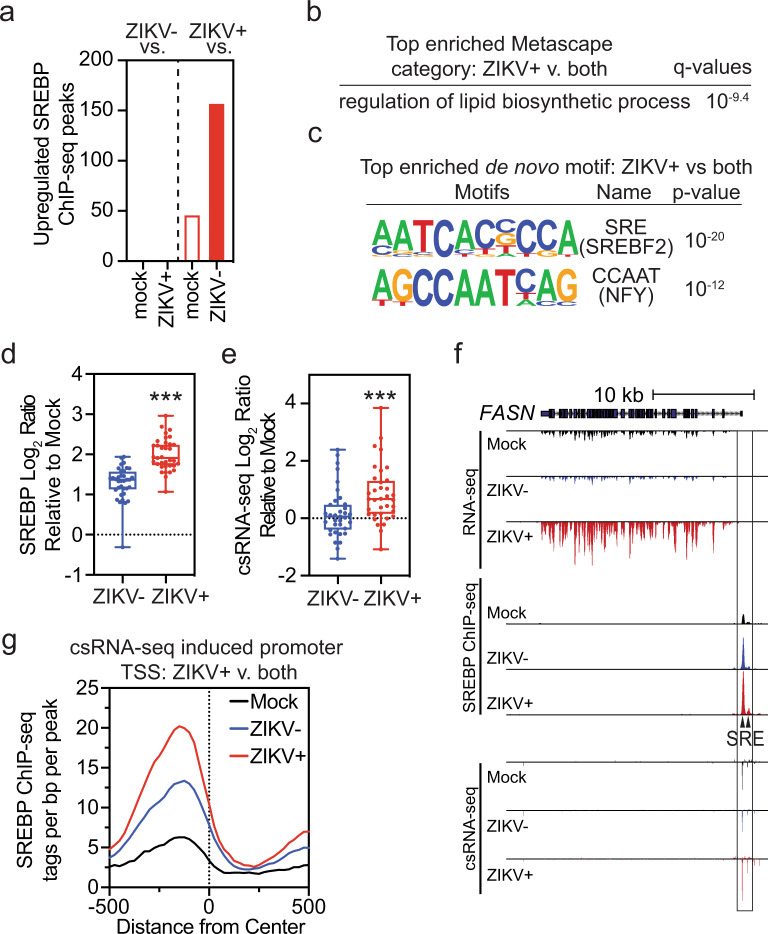
Fig. 4. ZIKV infection of human moDCs increases SREBP recruitment and transcription of lipid-related genes.
a–g ChIP-seq analysis was performed to detect SREBP TF binding to DNA in Mock, ZIKV−, or ZIKV+ moDCs from 3 donors infected for 24 h with ZIKV PRVABC59 (MOI 1). a Number of upregulated (fold-change >1.5, false discovery rate <0.1) SREBP peaks. b Most enriched Metascape pathways among genes with increased SREBP binding in ZIKV+ vs ZIKV− and Mock moDCs. c De novo motif analysis of upregulated SREBP peaks in ZIKV+ vs ZIKV− and Mock moDCs. d, e Log2 ratio of d SREBP or e csRNA-seq tags from −500 bp to +500 bp at upregulated SREBP peaks in ZIKV+ or ZIKV− vs Mock moDCs. All points representing upregulated ZIKV+ SREBP peaks are shown. Box plots show the median value and extend from the 25th to 75th percentile, while the whiskers extend from min to max values. N = 3 biologically independent experiments. ***P < 0.001 by two-sided paired t test. f UCSC browser visualization of the FASN locus with RNA-seq, SREBP ChIP-seq, and csRNA-seq in Mock, ZIKV−, and ZIKV+ moDCs. Location of SREBP (SRE) motifs is indicated. g SREBP-binding relative to csRNA-seq promoter proximal TSS upregulated in ZIKV+ vs ZIKV− and Mock moDCs. Source data and exact P values are provided as a Source Data file.