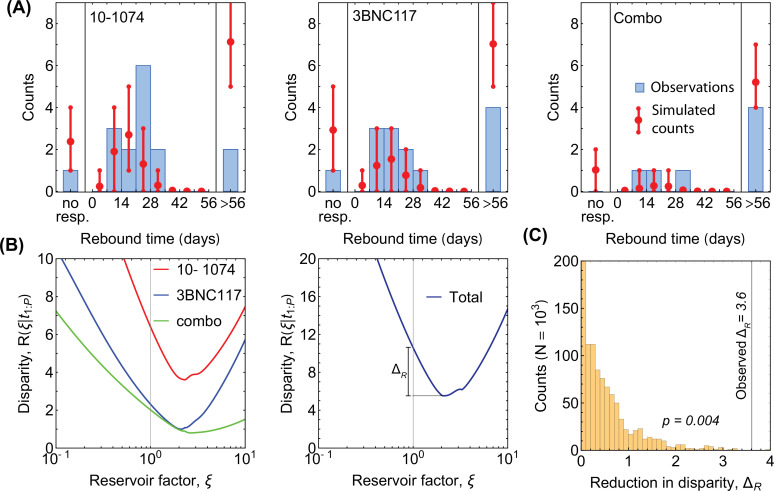
Figure 3. Statistics of viral rebound in clinical trials with bNAbs.
(A) Panels show viremia of three patients from the 3BNC117 trial over time (black circles) and the fitted model of the viral decay and rebound processes from Equation 1 (orange line). The viral rebound time and the fitted carrying capacity is shown in each panel. Shown are examples of a non-responder (NR; left), a rebound occurring during the trial window ( days; center), and a late rebound ( days; right). (B) We compare the distribution of rebound times in patients from the three clinical trials with 10–1074 Caskey et al., 2017, 3BNC117 Scheid et al., 2016, and the combination of the two bNAbs Baron et al., 2018 to the predictions from the simulations based on our evolutionary model (Figure 1, and Methods). The error bars show the inter decile range (0.1–0.9 quantiles) generated by the simulations for the corresponding trial. (C) The summary table shows the number of patients for whom the infecting HIV-1 population shows no response (NR), rebound during the trial window , and a late rebound ( days) in each trial. Note that three patients were excluded from the 3BNC117 trial (*) because of insufficient dosage leading to weak viral response: compared to the in the other treatment groups. (D) Plotted are 1,200 trajectories of the mutant viral population simulated using our individual based model. Due to the individual birth-death events, fluctuations are larger when the population size is smaller. At a critical threshold, , fluctuations are large enough to lead to almost certain extinction in the existing viral population. The critical threshold (yellow line) is an order of magnitude larger than the post-treatment spontaneously-generated mutant fraction (red line). (E) The predicted fraction of escape events associated with post-treatment spontaneous mutations (red) and the pre-treatment standing variation (yellow) are shown for the three trials. Late rebound events are indicated in blue. Because the spontaneously-generated mutant fraction is smaller than the extinction threshold, these mutations contribute to less than 4% of escape events (red), and escape is likely primarily driven driven by standing variation (yellow), that is, pre-existing escape variants.