FIG 1.
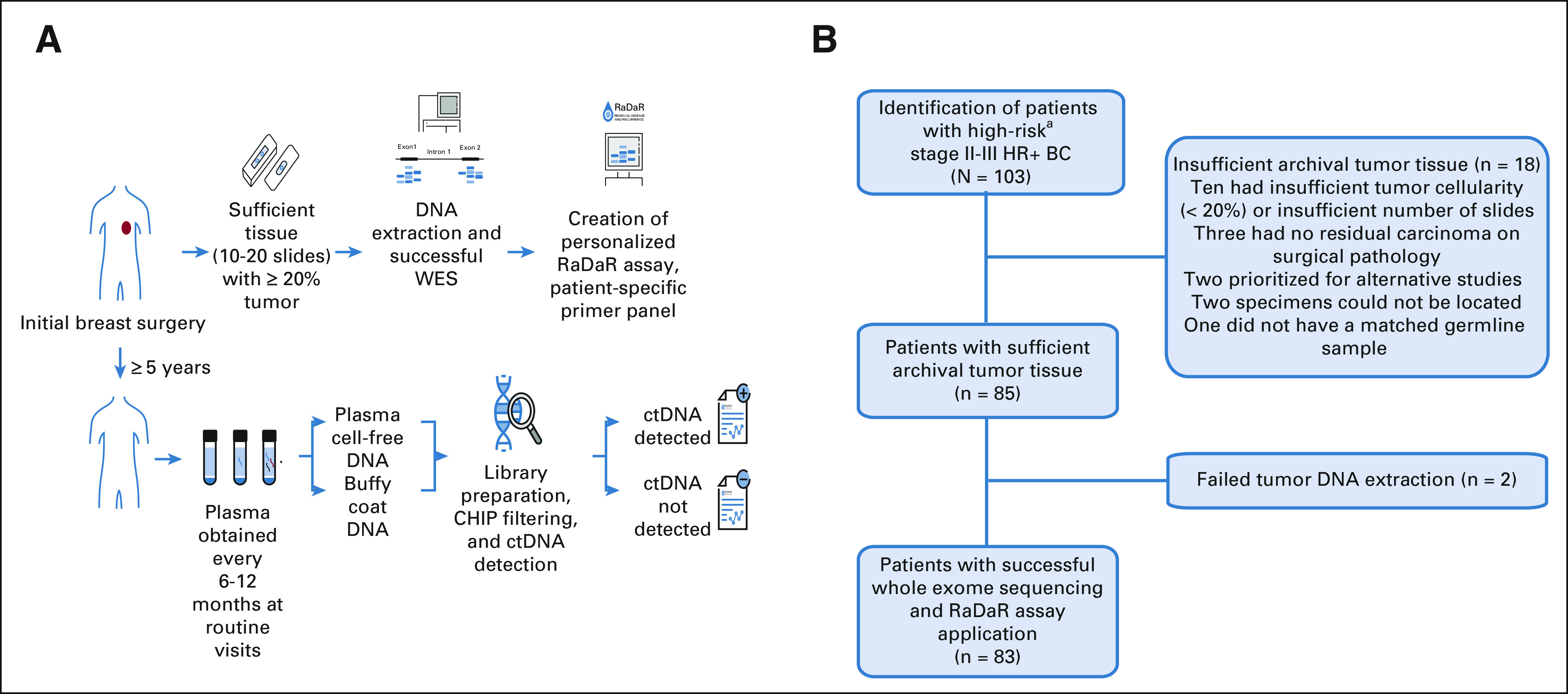
(A) RaDaR assay process. Primary tumor tissue from initial breast cancer surgery was obtained and assessed. For specimens with sufficient tissue (10-20 slides with 20% or higher tumor cellularity), DNA was extracted, and WES was performed to identify somatic mutations used to create personalized RaDaR assays for each patient. Patients were enrolled over 5 years after initial diagnosis, and every 6-12 months at routine follow-up visits, plasma was obtained, and the personalized RaDaR assay for ctDNA detection was applied. (B) CHiRP study flow chart. Patients with high-risk (T3/T4 and/or N2/N3; T1N1 and ≥ 3 involved lymph nodes; or T2N1 if: Ki-67 ≥ 20%, grade 3, or oncotype DX score ≥ 26) HR+ BC provided consent to obtain archival primary tumor tissue, and WES was performed. Patients were excluded from the analysis if there was insufficient primary tumor tissue or if WES was unsuccessful because of failed tumor DNA extraction. aHigh-risk: T3/T4 and/or N2/N3; T1N1 and ≥ 3 involved lymph nodes; or T2N1 if: Ki-67 ≥ 20%, grade 3, or oncotype DX score ≥ 26. CHIP, clonal hematopoiesis of indeterminate potential; CHiRP, circulating tumor DNA (ctDNA) and late recurrence in high-risk hormone receptor-positive, HER2-negative breast cancer; ctDNA, circulating tumor DNA; HR+ BC, hormone receptor–positive breast cancer; WES, whole-exome sequencing.
