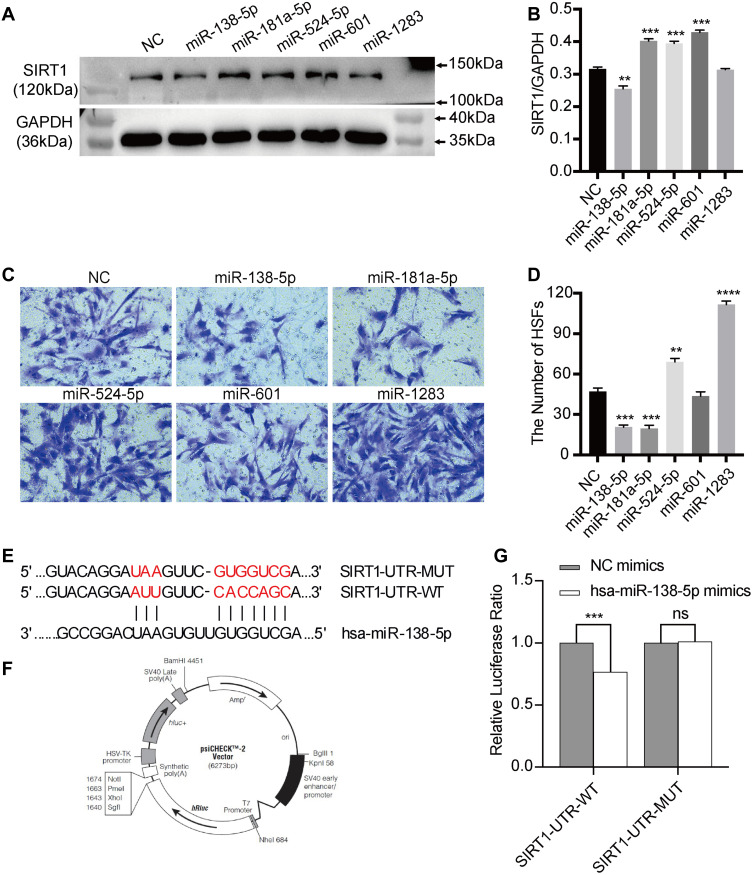
Figure 4.
The effects of miRNAs on protein expression of SIRT1 in HSFs and the migration of HSFs, and the verification of the targeted regulatory relationship between miR-138-5p and SIRT1 in HSFs. (A and B) Western blot analysis of protein expression of SIRT1 in HSFs exposed to miRNAs mimics or mimics NC for 48h, graph showed their levels relative to that of GAPDH. (C and D) Transwell migration analysis of the migration of HSFs exposed to miRNAs mimics or mimics NC for 48h, graph showed the numbers of migrated HSFs. (E) The binding sites predicted by bioinformatics algorithms. (F and G) The targeted modulation measured by luciferase reporter gene assays. Every experiment was repeated at least three times, and the data was shown as mean ± SEM (**P<0.01, ***P<0.001, ****P<0.0001, ns: no difference).
Abbreviations: SIRT1, silent information regulator 1; NC, negative control; HSFs, human skin fibroblasts; UTR, untranslated regions; WT, wild-type; MUT, mutant-type.