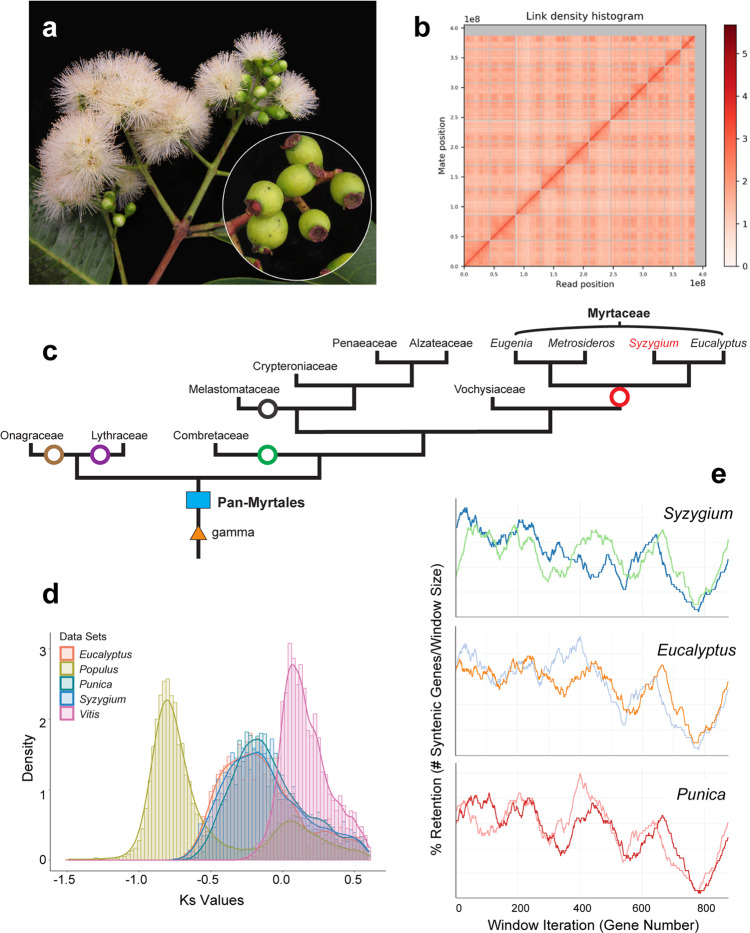
Fig. 1. Assembly and structural evolution of the Syzygium grande reference genome.
a S. grande inflorescence, flowers and fruits; the latter evoke the common name “sea apple”; b HiC contact map for the scaffolded genome, showing 11 assembled chromosomes; c Phylogeny of major lineages of Myrtales, following Maurin et al. 44. Genera of Myrtaceae used in genome structural and phylogenetic analyses are also depicted. Punica (Lythraceae) was also examined for structural evolution. Open circles represent the multiple, independent polyploidy events predicted by the 1KP study42; our results here suggest instead a single Pan-Myrtales whole genome duplication (blue rectangle) which followed the gamma hexaploidy (orange triangle) present in all core eudicots. d Synonymous substitution rate density plots for internal polyploid paralogs within Syzygium, Eucalyptus, Punica, Populus and Vitis. Modal peaks in these three Myrtales species suggest a single underlying polyploidy event. Ks asymmetries were calibrated using the gamma event present in each species. Both histograms and smoothed curves are shown. e Fractionation bias mappings of Myrtales chromosomal scaffolds, 2 each (different colours), onto Vitis vinifera chromosome 2 show similar patterns for all three Myrtales species (excluding cases of chromosomal rearrangements among the three, which are discernible as different scaffold colour switchings compared to the Vitis chromosome). X-axis shows the percent retention of fractionated gene pairs following polyploidisation; Y-axis shows the position of gene pairs along the Vitis chromosome. Photograph credit: WHL (a). Source data are provided as a Source Data file.