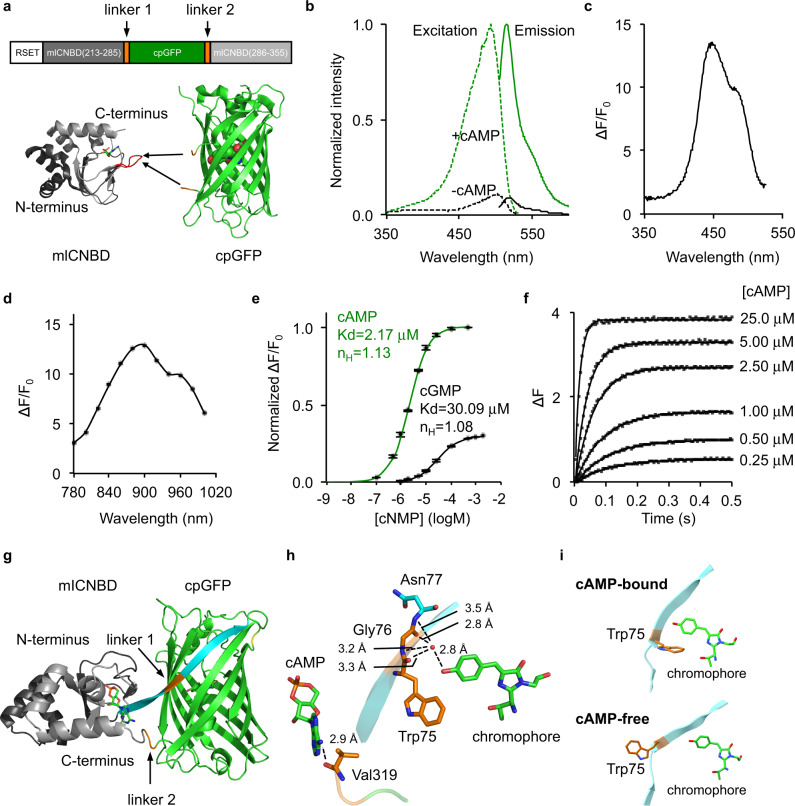
Fig. 1. Development and in vitro characterization of G-Flamp1 indicator.
a Schematic of G-Flamp sensors. cpGFP with two flanking linkers (two amino acids per linker) is inserted into mlCNBD (Gly213-Ala355, Genbank accession number: BA000012.4). The N-terminal peptide (RSET) including a 6× His tag is from the bacterial expression vector pNCS. The X-ray crystal structures of cAMP-bound mlCNBD (PDB: 1VP6) and cpGFP (PDB: 3WLD) are shown as cartoon with cAMP and chromophore of cpGFP shown as stick and sphere, respectively. The loop bearing the insertion site in G-Flamp1 is marked in red. b Excitation and emission spectra of cAMP-free and cAMP-bound G-Flamp1 sensors in HEPES buffer (pH 7.15). c Excitation wavelength-dependent ΔF/F0 of G-Flamp1 under one-photon excitation. d Excitation wavelength-dependent ΔF/F0 of G-Flamp1 under two-photon excitation. e Binding titration curves of G-Flamp1 to cAMP or cGMP in HEPES buffer (pH 7.15). The data were fitted by a sigmoidal binding function to extract the dissociation constant Kd and Hill coefficient nH. Data are presented as mean ± standard error of mean (SEM) from three independent experiments. f Binding kinetics of G-Flamp1 to cAMP measured using the stopped-flow technique in HEPES buffer (pH 7.15). Each curve corresponds to a different concentration of cAMP, i.e., from bottom to top: 0.25, 0.5, 1, 2.5, 5, and 25 μM. The data were fitted by a single-exponential function. g Cartoon representation of crystal structure of cAMP-bound G-Flamp1 (PDB: 6M63). The N- and C-terminal fragments of mlCNBD are shown in dark and light gray, respectively. cpGFP is in green and both linkers are in orange. The long β-strand possessing linker 1 is in cyan. h Chromophore and cAMP are in close proximity with linker 1 and linker 2, respectively. i Zoom-in view of Trp75 and the chromophore of simulated cAMP-bound and cAMP-free structures at conformations associated with the global minimum energy. Source data are provided as a Source Data file.