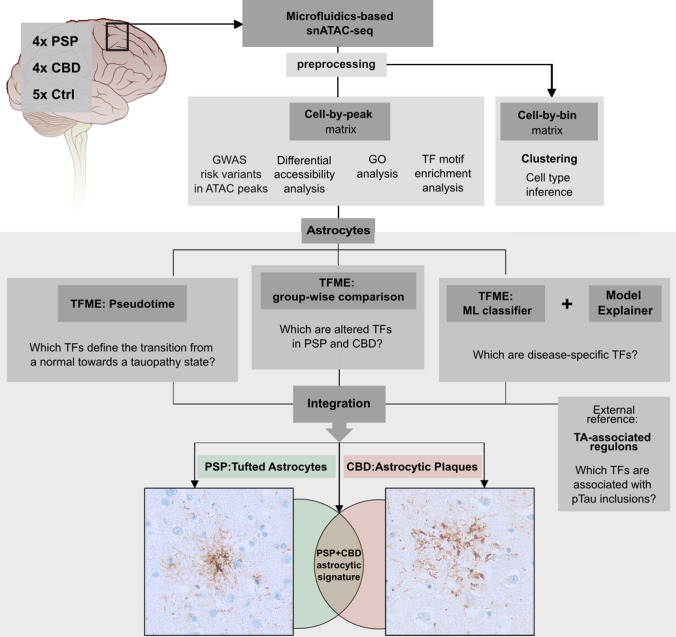
Fig. 1.
Concept of the bioinformatical analysis. SnATAC-sequencing was applied to snap-frozen frontal cortex samples from deceased PSP, CBD, and Ctrl individuals. Raw sequencing reads were preprocessed and resulting matrices were then used (i) for graph-based clustering and cell type inference (using a binned genome), and (ii) for GWAS risk variant-association with cell types, differential accessibility analysis and GO, as well as TF-motif analysis (using the peak matrix). Downstream, only the astrocytic cluster was investigated (boxed lower part). To find significantly altered TFs in tauopathy-derived astrocytes, disease-wise comparisons of TFME were conducted (mid panel). TFME changes along pseudotime trajectories were assessed to identify TFs linked with pathogenesis (left). An ML-based disease classifier was utilized to delineate disease-specific TFs in a more unbiased approach (right). Significant results from these three branches were refined by a TF profile linked to the presence of astrocytic pTau inclusions in PSP (Tufted Astrocytes, TA). Finally, this multilayered regulon pattern was integrated to define a general astrocytic tauopathy TF signature, or entity-specific astrocytic TF signatures. These are presumed to mirror the neuropathological context of characteristic pTau inclusions in astrocytes, namely TA in PSP and AP in CBD. AP astrocytic plaque, GO gene ontology, GWAS genome wide association studies, ML machine learning, pTau hyperphosphorylated Tau, TA tufted astrocyte, TFME transcription factor motif enrichment. The brain illustration was modified from https://de.m.wikipedia.org/wiki/Datei:Brain_stem_normal_human.svg (CC-Attribution-2.5 License 2006)