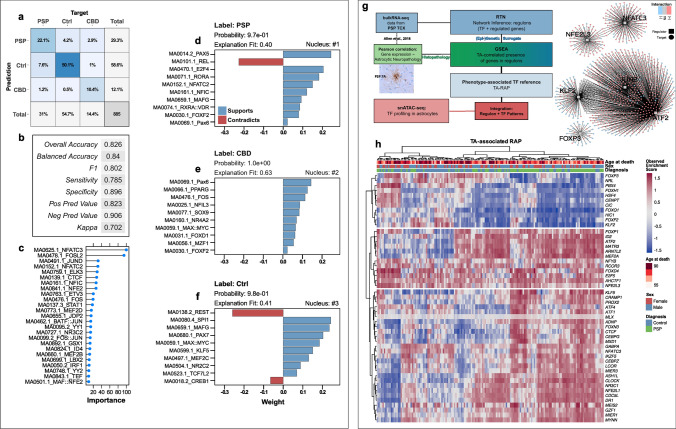
Fig. 5.
TF networks associated with the astrocytic tauopathy state and regulatory correlates of tufted astrocytes. a Confusion matrix displaying the intersections of the XGB model’s predictions (rows) and the actual labels (columns). Each square contains the percentual proportion of test set samples (= 20% nuclei) with the assigned prediction–label relation. The sums of each row or column are depicted in the rightmost column or bottom row, respectively. The total sample number (i.e., nuclei of the 20% test set-split) is shown in the bottom right corner. b Evaluation parameters of classification performance of the trained XGB model on the 20% test set-split. Overall, more than 82% of predictions were correct (overall accuracy) and the model performs "substantially" with a Cohen kappa of 70.2%. c Overall feature importance values of the top 25 TFMs included in training the XGB model to correctly classify an astrocyte TF representation in general. The x-axis differentiates the feature importance (%) as reported by caret’s varImp function. Immediate-early response candidates (NFAT2/3) and major AP-1 constituents (FOSL2, JUND) were among the most important TFs. d–f Lime feature importance bar diagrams of the most certainly correctly classified barcodes of each group entity. The bar direction and bar color indicate the feature weights (~ importance) assigned to the TFM, which are given as y-axis breaks. Feature weight was assigned to specific TFME value ranges. Each panel is complemented by the group entity label, the model's calculated probability, and the explanatory model’s fit value. g Bioinformatical concept of the RTN analytical approach to link a neuropathological phenotype to TF information. A regulon network was inferred from published bulkRNA-seq data in PSP TCX and filtered subsequently for those regulons that showed phenotype association (i.e., gene set enrichment of DEGs with histopathological TA grading in PSP cortices). Thereby, a TA-associated regulon activity profile was deduced, which was employed as TF reference in an integration part with snATAC-seq data-derived astrocytic TF activity patterns. Ultimately, this approach served to refine pTau-inclusion pathology-associated astrocytic PSP/CBD signatures. On the right, a set of TA-linked regulons illustrates the modularity of TF-gene-interactions (color code), the inter-modular connectivity suggesting co-regulation exerted by regulators on common genes, and the presumed presence of distinct groups of TFs. h Activity heatmap of those regulons that are enriched with TA grading in PSP TCX and whose regulon activity is significantly different between PSP and Ctrl TCX samples (p < .05, BH-corrected). Regulons in the upper part correlate negatively, those in the lower part correlate positively with TAs in PSP cortices. Every column corresponds to a single TCX sample and every row to a gene while color shade indicates the extent of regulon activity change. Rows and columns were clustered hierarchically (Euclidean distance, Ward-D2 method) and results indicated as dendrograms. The colored overlay informs about the age at death, sex, and definitive neuropathological diagnosis. Gene names comply with the Ensembl IDs. DEG differentially expressed gene, GSEA gene set enrichment analysis, Neg Pred Value negative predictive value, Pos Pred Value positive predictive value, RTN Reconstruction of Transcriptional Networks, TA tufted astrocyte, TCX temporal cortex, RAP regulon activity profile, XGB extreme gradient boosting tree