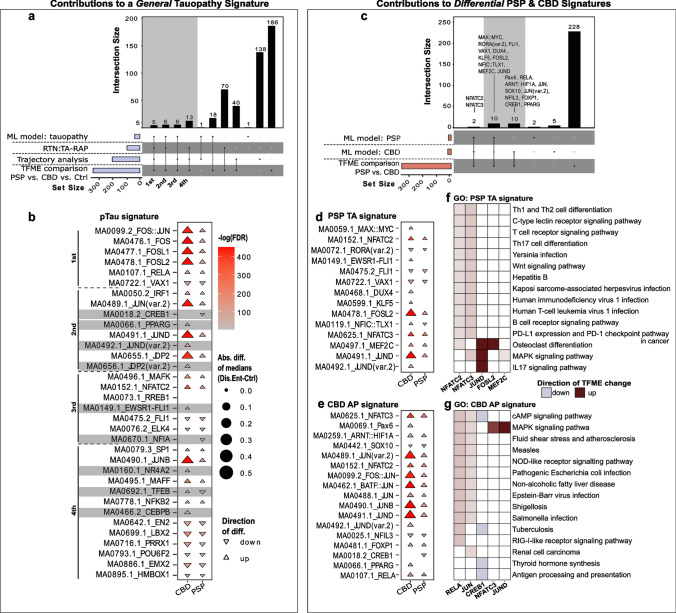
Fig. 6.
A concept of astrocytic tauopathy signatures. a Upset plot illustrating TFs useful in distinguishing PSP/CBD from Ctrl astrocytes that resulted from (i) interpreting the XGB classification model (‘ML model: tauopathy’), (ii) the TA-related regulon activity profile in the bulkRNA-seq data set (‘RTN: TA-RAP’), (iii) the pseudotime trajectory analysis in the snATAC-seq data set (‘Trajectory analysis’), and (iv) group-wise TFME comparisons in the snATAC-seq data set (‘TFME’ comparison’). Set sizes are indicated as blue bars, while the intersection logic is shown as vertical lines and dots. Column heights depict the extent of intersection for the given sets. The first four intersections were assigned a hierarchy of importance in defining the primary tauopathy context. b Triangle plot indicating significance, absolute extent, and direction of TFME changes in pTau signature TFs in tauopathy-assigned astrocytes. The triangle tips point towards the direction of change while the size represents the absolute difference from the TFME reference in Ctrls. Fill shading displays the negative decadic logarithm of the BH-corrected p values from pair-wise Wilcoxon rank-sum tests. Empty coordinates inform about non-significant comparisons. Gray underlay informs about candidates with diverging TFME when collating PSP and CBD. c Upset plot to identify TFs useful in differentiating CBD from PSP astrocytes. The single sets resulted (i) from the most important TFs for PSP or CBD prediction according to the XGB model explainer (‘ML model: PSP’, ‘ML model: CBD’) and (ii) from pairwise statistical TFME comparisons between PSP and CBD astrocytes (‘TFME comparisons’). The general plot structure equates to A. d, e Triangle plot indicating significance, absolute extent, and direction of TFME changes in PSP (d), and CBD (e) signature TFs in tauopathy-assigned astrocytes. The general plot structure equates to B. f, g Heatmaps of the GO enrichment of the PSP (f) and CBD TF signatures (g). The top 15 terms according to MF, BP, and CC enrichment scores as well as only those TFs that share at least one of these terms are depicted. Color code indicates the direction and strength of enrichment or depletion compared to Ctrl astrocytes. MAPK signaling, immunological and infectious disease terms are enriched. Abs.diff. absolute difference, AP astrocytic plaque, CMA chaperon-mediated autophagy, DAR differentially accessible region, Dis.Ent. disease entity, EC extracellular, Exc. DLN excitatory deep-layer neurons, Exc. ULN excitatory upper-layer neurons, FDR false discovery rate, GO gene ontology, Mic microglia, ML machine learning, Oli oligodendrocytes, OPC oligodendrocytic precursor cells, TA-RAP tufted astrocyte-associated regulon activity profile, TF(ME) transcription factor (motif enrichment), UPS ubiquitin–proteasome-system, UPR unfolded-protein-response