Fig. 7.
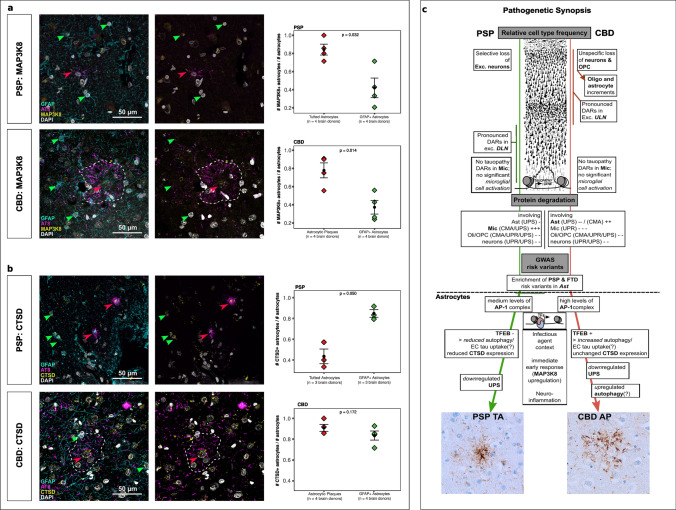
TF target gene validation and synopsis of pathogenesis. a Immunofluorescent staining analysis showing 4-channels merge (GFAP, AT8, MAP3K8, DAPI) and 3-channels merge (AT8, MAP3K8, DAPI) of PSP (upper row) and CBD (lower row) MFG. Arrowheads mark GFAP + (green) and AT8 + (red) astrocytes. Boxplots depicting fractions of GFAP + MAP3K8 + astrocytes over GFAP + astrocytes compared with the fractions of AT8 + MAP3K8 + astrocytes over AT8 + astrocytes. Statistics were calculated using a two-tailed paired t-test with p values as indicated. b Immunofluorescent staining analysis showing 4-channels merge (GFAP, AT8, CTSD, DAPI) and 3-channels merge (AT8, CTSD, DAPI) of PSP (upper row) and CBD (lower row) MFG. Arrowheads mark GFAP + (green) and AT8 + (red) astrocytes. Boxplots depicting fractions of GFAP + CTSD + astrocytes over GFAP + astrocytes compared with the fractions of AT8 + CTSD + astrocytes over AT8 + astrocytes. Plot structure equal to a. c Concept of epigenetic contribution to the pathogenesis in the primary 4R tauopathies PSP and CBD. The upper half summarizes global findings of this study, while the lower half focuses on changes assigned to astrocytes. Differences in neuronal cell loss were observed and mirrored by prominent DAR-patterns in different neuronal subclusters. Protein degradation was induced in Mic in PSP, while Ast served this role in CBD. PSP and FTD-associated risk variants were exclusively enriched in Ast. Focusing on the latter glia type, disease-specific molecular patterns comprising regulators of the immediate early response, autophagy, and UPS delineate differential pathogenetic signatures. The histological illustration of the neocortex was modified from https://commons.wikimedia.org/wiki/File:Cajal_cortex_drawings.png. AP astrocytic plaque, CMA chaperon-mediated autophagy, CTSD Cathepsin D, DAR differentially accessible region, Dis.Ent. disease entity, EC extracellular, Exc. DLN excitatory deep-layer neurons, Exc. ULN excitatory upper-layer neurons, FDR false discovery rate, GO gene ontology, MAP3K8 Mitogen-activated protein 3 kinase 8, Mic microglia, ML machine learning, Oli oligodendrocytes, OPC oligodendrocytic precursor cells, TA-RAP tufted astrocyte-associated regulon activity profile, TF(ME) transcription factor (motif enrichment), UPS ubiquitin–proteasome-system, UPR unfolded-protein-response