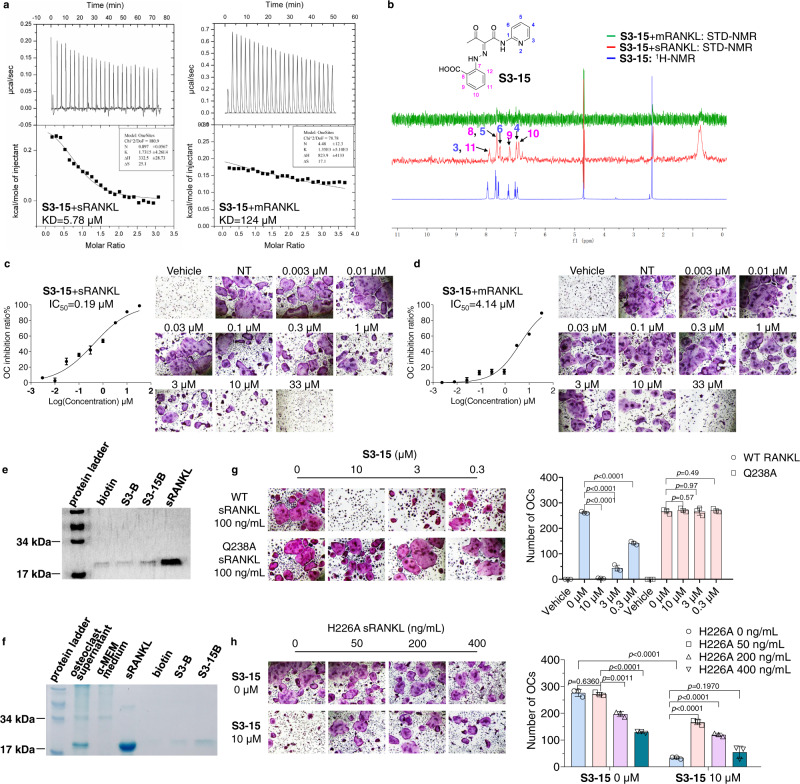
Fig. 3. S3-15 selectivity binding to sRANKL.
a Binding of S3-15 to sRANKL or L-RANKL as measured by ITC. b Ligand-observed 1D 1H-NMR of the aromatic peaks of S3-15 (blue) shows chemical shift changes in buffer. 1H-NMR STD transfer experiments show the appearance of aromatic peaks of S3-15 in the presence of sRANKL (red color), while the aromatic peaks disappear in the presence of L-RANKL (green) because of the low binding affinity of S3-15 to mRANKL. In the structure of S3-15, the 1H-NMR aromatic peaks locate in 3, 4, 5, 6 position in the pyridine ring and 8, 9, 10, 11 position in the benzene ring. c, d S3-15 inhibits sRANKL induced osteoclast differentiation, while exhibits weak osteoclast differentiation in mRANKL induced osteoclastogenesis assay. The picture shows the IC50 curve and a representative picture of each concentration. Scale bars, 75 μm. e Pull down assays show that S3-15B can bind to sRANKL, while S3B showed no binding with sRANKL. ~f S3-15B selectively bind to sRANKL over other proteins in supernatant of osteoclast differentiation culture. g S3-15 is failure to bind sRANKL Q238A mutant, and the osteoclastogenesis is not inhibited. h sRANKL H226A mutant can partially bind with S3-15 but cannot induce osteoclast, S3-15 still able to inhibit osteoclastogenesis even S3-15 is partially occupied when stimulated with 100 ng/mL WT sRANKL and different concentrition of H226A sRANKL. (Left, OC TRAP-stained pictures taken from one of three biological replicates, scale bars, 75 μm. Right, the OC numbers were counted in each well and presented in mean± s.d.) Statistical difference was determined by unpaired two-tailed Student’s t test. Significant difference p value is < 0.05.