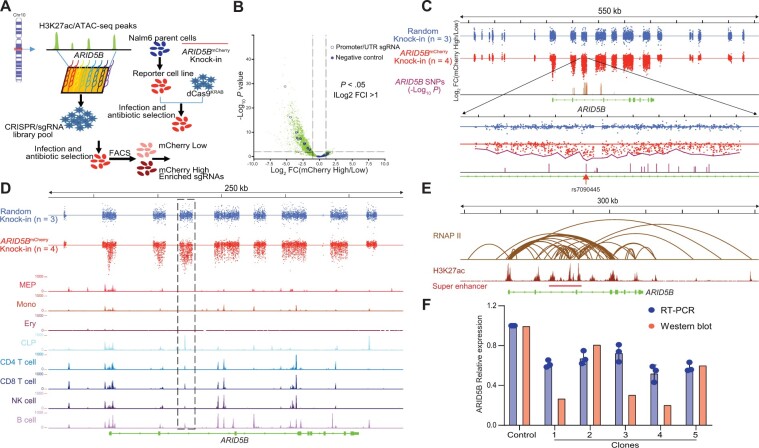
Figure 2.
CRISPR/dCas9-KRAB library screening for the interrogation of CREs of ARID5B. A) An overall schema of the design strategy. A single-guide RNA (sgRNA) library was designed using GM12878 H3K27ac chromatin immunoprecipitation sequencing (ChIP-seq) data and normal human hematopoietic cell assay of transposase accessible chromatin sequencing (ATAC-seq) data for reference. A single-cell clone reporter cell line stably expressing dCas9-KRAB was established then infected with CRISPR/sgRNA library pool virus at low multiplicity of infection (0.2) to ensure that each cell expressed no more than 1 sgRNA. Flow sorting was used to differentiate the mCherry-low population (the bottom 10%) and the mCherry-high population (the top 10%). Genomic DNA from these 2 populations was then sequenced to calculate the enrichment of each sgRNA. B) A volcano plot was generated to show the enrichment of sgRNAs in the mCherry-high or the mCherry-low populations. In total, 10 497 sgRNAs were designed, including 10 sgRNAs (open blue circles) that target the ARID5 B’s promoter and untranslated region, and 10 negative sgRNAs (solid blue circles) that do not target any location in the human genome. P values of .05 and |log2 FC| = 1 were used as cutoffs. C) The CRISPR/dCas9-KRAB library screening results. A Nalm6 single-cell clone with random knock-in mCherry was used as a negative control (top). For ARID5B mCherry knock-in cells (middle), sgRNAs were overrepresented mainly in predicted enhancers, including the ARID5B promoter, and were less represented in other regions. In contrast, for the negative control, sgRNAs were distributed almost equally across the whole ARID5B gene (data were derived in triplicate). Six cis-regulatory elements (CREs) were nominated out of 21 segments targeted by the sgRNA library from upstream to downstream of the ARID5B gene by 2-sided t test (P < .001) via comparing each segment of ARID5BmCherry knock-in with that of the random knock-in. The highlighted region shown encompasses SNPs having the strongest associations with ALL. In particular, SNP rs7090445 is juxtaposed with the most highly enriched sgRNA. D) Alignment of CRISPRi screening results with human hematopoietic cells ATAC-seq data. The most statistically significant CRE overlapped with lymphoid lineages (common lymphoid progenitors, CD4 T cell, CD8 T cell, natural killer cell, and B cell) specific ATAC-seq peaks (highlighted in the dashed box). E) A public GM12878 chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) dataset was used to interrogate the CRISPR/dCas9-KRAB library data. There were strong interactions between the ARID5B promoter and CREs nominated based on CRISPRi screening, as shown by the DNA loop generated from RNA polymerase II ChI A-PET data. The most statistically significant CRE is located in a super enhancer. F) Part of the highlighted region (chr10:61 960 755–61 962 481, 1.7 kb) in panel C was deleted using CRISPR/Cas9, resulting in a statistically significant decrease in ARID5B expression at both the protein and mRNA levels throughout all 5 single-cell clones.