Highlights
-
•
Circ-0010117 is down-regulated in glioblastoma.
-
•
Circ-0010117 regulates aggressiveness via miRNA-6779-5p in glioblastoma.
-
•
SPEN contributed to regulate miRNA-6779-5p in glioblastoma.
-
•
Upregulated Circ-0010117 inhibited in vivo tumor growth of human glioblastoma xenograft.
Keywords: Glioblastoma, circ-0010117, miR-6779-5p, SPEN
Abstract
Noncoding RNAs (ncRNAs) play important roles in cancer biology, providing potential targets for cancer intervention. As a new class of endogenous noncoding RNAs, circular RNAs (circRNAs) have been recently identified in cell development and function, and certain types of pathological responses contribute to cancer progression, including glioblastoma. However, the potential mechanisms underlying the relationship between circRNAs and glioblastoma progression are still largely unknown. Methods: The expression and roles of circular RNA 0010117 (circ-0010117) were examined in vitro and in vivo. Quantitative RT‒PCR and western blotting were used to measure the expression of circRNA, miRNA, each gene, or related proteins. Cell biology experiments were performed to detect the biological function of circ-0010117 in glioblastoma cell lines. Moreover, bioinformatics analysis, luciferase reporter assays, and functional complementation analysis were carried out to investigate the target genes. Tumorigenesis was also evaluated by xenografting cells into nude mice. In this study, we found that circ-0010117 is downregulated in glioblastoma compared with corresponding paratumoural tissues. Subsequently, we observed that circ-0010117 can regulate aggressiveness in glioblastoma cells through miR-6779-5p. Furthermore, SPEN was verified as a direct target of miR-6779-5p and contributes to the circ-0010117 regulatory network. In addition, we identified that overexpression of circ-0010117 can suppress tumorigenesis in nude mice. Our findings indicate that circular RNA 0010117 promotes the aggressive behavior of glioblastoma by regulating the miRNA-6779-5p/SPEN axis. Our results provide a rationale for the use of circ-0010117 as a novel potential therapeutic target in glioblastoma.
Introduction
Glioblastoma is the most common primary brain cancer of glial origin worldwide and is the leading cause of neurologic cancer-related death worldwide [1]. High-grade glioblastoma in particular has a very poor prognosis and poses a serious threat to human health. In recent years, although advances have been made in surgery, chemotherapy and molecular targeted therapies, glioblastoma still has poor morbidity and mortality. Given the poor strategies for diagnosis and treatment, a better understanding of glioblastoma pathogenesis is critical for advancing and providing potential biomarker-driven treatments.
Noncoding RNAs (ncRNAs) are a class of nonprotein translation RNAs that include long noncoding RNAs (lncRNAs) more than 200 nucleotides in length, microRNAs (miRNAs) 20–22 nucleotides in length, and circular RNAs (circRNAs), which are characterized by covalent closed loops. Similar to other ncRNAs, circRNAs are expressed in multiple tissues with great variability. CircRNAs are a special class of noncoding RNAs that originate from exons, introns, intergenic regions or untranslated regions by a noncanonical splicing process termed back splicing [2]. Importantly, circRNAs participate in the occurrence of many kinds of tumors and regulate tumor development. Circular RNA hsa_circ_0001785 has been reported to inhibit the proliferation, migration and invasion of breast cancer cells in vitro and in vivo by sponging miR-942 to upregulate SOCS3 (suppressors of cytokine signaling 3) [3]. The circular RNA hsa_circ_0079,929 has also been validated to inhibit tumor growth in hepatocellular carcinoma [4]. Another novel circular RNA, circENTPD7, has been confirmed to contribute to glioblastoma progression by targeting ROS1 (ROS proto-oncogene 1) [5]. However, the potentially important functional role of circ-0010117 in the progression of glioblastoma is still unclear.
In this study, we first investigated the expression of circ-0010117 in human glioblastoma tissues. We subsequently investigated the effects of circ-0010117 on the aggressiveness of human glioblastoma cells. Furthermore, we also explored the molecular mechanisms underlying these effects.
Materials and methods
Human osteosarcoma clinical specimens
A total of 6 paired osteosarcoma and corresponding paratumoural tissues were collected at the First Affiliated Hospital of Nanchang University from November 2021 to January 2022. All tissue specimens were resected during surgery and directly snap frozen in liquid nitrogen. The study protocol was approved by the ethics committee of the First Affiliated Hospital of Nanchang University. All written informed consent was received from patients who underwent surgical resection.
Cell culture
The U251 and U87 cell lines (procell, Wuhan, China) are commonly used as experimental models of glioblastoma. Cells were cultured in Dulbecco's modified Eagle's medium (Gibco, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, Waltham, MA, USA), pyruvate (1 mM; Sigma‒Aldrich, Shanghai, China), nonessential amino acids (0.1 mM; Sigma‒Aldrich, Shanghai, China), and a mix of antibiotics (1 mM; Sigma‒Aldrich, Shanghai, China) in a humidified atmosphere with 5% CO2 at 37 °C.
Quantitative RT-PCR
Total RNA from tissue samples and cells was isolated using the mirVanaTM miRNA isolation kit (Ambion, Austin, TX, USA) according to the manufacturer's instructions. RT‒qPCR analysis was performed with SYBR green in a 7500 Fast Real-Time PCR system (Applied Biosystems, Carlsbad, CA, USA). Circ-0010117, microRNA-6779-5p (miR-669-5p) and SPEN were measured by qRT‒PCR. U6 was used as an internal reference for circ-0010117 and miR-669-5p, as previously described [6]. β-Actin was used as an internal control for SPEN. All primers were commercially available from Sangon Biotech (Shanghai, China). The amount of circRNA, miRNA or each gene relative to the internal control was calculated as previously described [7].
RNA interference of circ-0010117
Circ-0010117 siRNAs (siRNA-#1, siRNA-#2 and siRNA-#3) targeting the junction region of the circRNA-0010117 sequence were commercially available from GeneCopoeia (Rockville, MD, USA). Transfection of siRNA was performed using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. shcirc-0010117 is also commercially available from GeneCopoeia (Rockville, MD).
For the circ-0010117 overexpression plasmid (circ-0010117), the sequence was cloned into GV248. The backbone of pLCDH-ciR (Geneseed, Guangzhou, China) was used to construct the circ-0010117 overexpression plasmid. The linear sequence of circ-0010117 together with the flanking circ-0010117-inducing sequence was amplified and subcloned into the EcoRI and BamHI sites of pLCDH-ciR.
MiR-6779-5p/SPEN overexpression and inhibition
For SPEN overexpression in vitro, we used an SPEN vector construct (OriGene Technologies, Inc., Rockville, MD). U251 and U87 cells were seeded in a 6-well plate (Corning, Cambridge, MA, USA) (2 × 105 cells/well) with complete medium. The cells were transfected with 2 μg of SPEN vector or the vector control (OriGene Technologies) using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) with Opti-MEM (Gibco) for 4 h. Then, we replaced the Opti-MEM with complete medium. After 48 h, the cells were collected for the next determination. MiR-6779-5p mimics are commercially available from life technologies. Transfection of miRNA mimics was performed using RNAiMAX Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. miRNA vector control (NC mimic) was used as a vector control. The shSPEN and miR-6779-5p inhibitor are also commercially available from GeneCopoeia (Rockville, MD).
Cell Counting Kit-8 (CCK8) assay
A CCK-8 assay was performed to assess the proliferation of U251 and U87 cells. For the CCK-8 assay, mock and infected cells (1 × 105 cells/well) were seeded into 96-well plates (Corning, Cambridge, MA, USA) after 24 h of transfection and cultured for 24 h, 48 h and 72 h. The absorbance at 450 nm was measured using CCK-8 (Dojindo Laboratories, Kumamoto, Japan) at the indicated times according to the manufacturer's protocols.
Colony formation assay
For the colony formation assay, we melted 1.4% (v/v) agarose (Sigma‒Aldrich, Shanghai, China) in a microwave and cooled it to room temperature. Then, we mixed equal volumes of the 1.4% melted agarose with the complete cell culture medium. After that, 2 ml of 0.7% (v/v) low melt agar was added into each well of the 6-well plate (Corning, Cambridge, MA, USA) and set aside to allow agarose to solidify. Cells (5 × 103 cells/well) were mixed with 1.4% agarose in complete culture medium and plated on top of the solidified layer to form colonies at 1–3 weeks. Cells were fed complete culture medium every 3 d.
The colony is required to consist of at least 50 cells [8]. Clones were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet (Millipore, Billerica, MA, USA). Visible colonies were manually counted.
Apoptosis assay
Flow cytometry was performed for the apoptosis analysis. U251 and U87 cells were digested with trypsin (Invitrogen, Carlsbad, CA, USA) and washed with PBS. After blocking with 10% FBS, the cells were stained with 5 μl FITC annexin-V and 1 μl propidium iodide (Becton Dickinson, Heidelberg, Germany) and incubated at room temperature for 15 min in the dark. Then, flow cytometry was performed (Life Technologies, Darmstadt, Germany). The cells were then analyzed with CellQuest software (BD Biosciences, San Jose, CA). The dead cells and the relative ratio of apoptotic cells were compared in each condition.
Wound scratch assay analysis
U251 and U87 cells were first transfected with shcirc-0010117 or circ-0010117 or their related control for 24 h. A wound scratch assay was performed as previously described [9], [10], [11], [12]. Then, the cells were seeded into 12-well plates (5 × 105 cells/well) with medium and cultured for 24 h to reach ∼90% confluence. A 10 μl pipette tip was used to make a scratch line. The cells were washed with PBS 3 times to remove the scratched cells. Then, the cells were cultured in medium containing 0.5% FBS. The cells were incubated at 37 °C, and images were captured at 0 and 48 h using a light microscope (Olympus Corporation, Tokyo, Japan).
Cellular invasion
A Boyden chamber assay with a 24-well collagen-based cell invasion assay kit (Millipore, Bedford, MA, USA) was used to perform the transwell assay. U251 and U87 cells were first transfected with shcirc-0010117 or circ-0010117 or their related control for 24 h. Then, 200 μl of serum-free medium containing 5 × 105 cells was added to the filter. Then, 750 μl of complete cell culture medium was added to the bottom chamber, in which FBS was used as a chemoattractant. After incubation for the indicated time, the noninvasive cells were cleaned by scrubbing with a cotton swab. The cells that adhered to the outside of the membrane were fixed and dyed with Cristal Violet solution (Millipore, Billerica, MA, USA). An Olympus optical microscope (Olympus Corporation, Tokyo, Japan) was used to observe invasive cells.
Dual-luciferase reporter assay
The binding sites of circ-0010117 for miR-6779-5p of its 3’-UTR and mutant were commercially available at GeneCopoeia (Rockville, MD). U251 and U87 cells were seeded in a 6-well plate (Corning, Cambridge, MA, USA) at a density of 2.5 × 105 cells/well with complete medium. The cells were cotransfected with 1 µg circ-0010117 3’-UTR (circ0010117-WT) or mutant luciferase reporter construct (circ0010117-Mut) with 20 nM miRNA mimic or NC mimic using Lipofectamine 2000 (Carlsbad, CA, USA) with Opti-MEM (Gibco, Waltham, MA, USA) for 48 h. Luciferase assays were performed using the dual-luciferase reporter assay reagent from GeneCopoeia. Data are expressed as the ratio of firefly luciferase activity to Renilla luciferase activity.
Western blotting
Total protein was extracted from cells with and without circ-0010117 with T-PER Tissue Protein Extraction Reagent (Thermo Pierce, Rockford, IL, USA) with 1% Halt Protease and Phosphatase Inhibitor (Thermo Pierce, Rockford, IL, USA). Then, the extracted protein was loaded and run on sodium dodecyl sulfate-polyacrylamide gels (8–12% separating gel, 5% concentrated gel). The protein in the gel was transferred onto a PVDF membrane (Millipore, Bedford, MA, USA) for antibody detection. The PVDF membrane was blocked for 1 h at room temperature in 5% nonfat milk in Tris-buffered saline with 0.1% Tween-20 detergent (TBST). The membrane was washed with TBST 3 times for 10 min each at room temperature and incubated with primary antibodies (anti-truncated-cas-3, full-cas-3, cas-8, cas-9, BCL-2, BAX, P53, and JNK1) overnight at 4 °C. β-actin served as an internal control (Abcam, Cambridge, MA, USA). Then, the membrane was washed with TBST 3 times for 10 min each at room temperature and incubated with horseradish peroxidase (HRP)-labeled goat anti-mouse IgG or goat anti-rabbit IgG secondary antibody (Thermo Pierce, Rockford, IL, USA) for 1 h at room temperature.wAfter the membrane was washed with TBST 3 times for 10 min each at room temperature, proteins were visualized by using ECL western blotting detection reagents (SuperSignal® West Dura Extended Duration Substrate, Thermo Pierce, Rockford, IL, USA). The relative expression of the target protein was quantitatively analyzed by ImageJ (NIH, Bethesda, MD, USA).
Mouse xenograft assays
Female nude mice (BALB/c, SPF grade, weighing 16–20 g and 4 weeks old) were purchased from Shanghai Lingchang Biotechnology Co., Ltd. (Shanghai, China). Transfected cells were digested with trypsin and washed twice with PBS. Cells were resuspended in 200 µL solution containing 100 µL PBS and 100 µL Matrigel substrate at a final density of 2 × 107 cells/0.2 mL. Four-week-old female BALB/c nude mice were subcutaneously injected with 2 × 107 tumor cells. Mice were sacrificed at Day 35 after tumor cells had been injected, and tumor volume was measured.
Statistical analysis
Statistical analyzes were performed using GraphPad Prism, and all data are presented as the mean ± standard deviation (SD) from triplicate experiments. Differences between groups were analyzed by unpaired t test. A p value < 0.05 was considered significant.
Results
Circ-0010117 is downregulated in glioblastoma
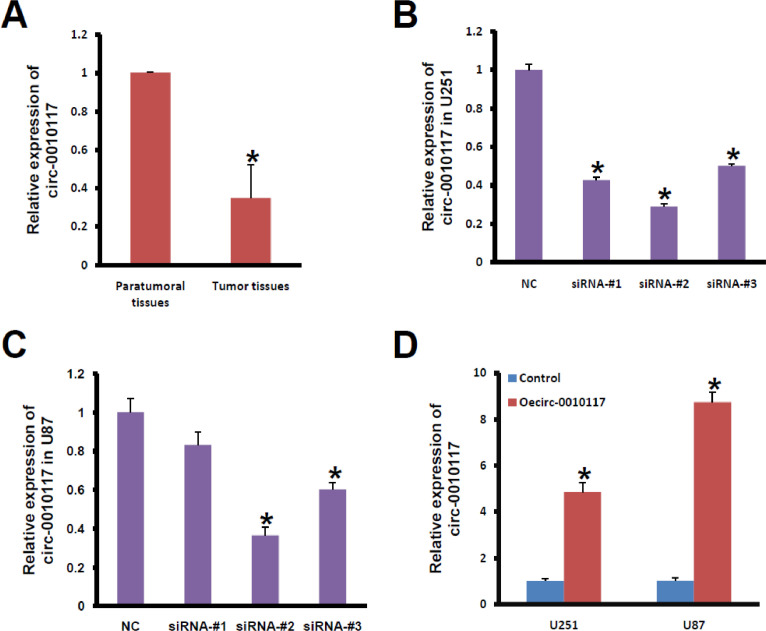
In this study, we first determined the mRNA expression levels of circ-0010117 in 6 paired glioblastoma and corresponding paratumoural tissues with qRT‒PCR. Circ-0010117 mRNA levels were significantly downregulated compared with those in normal tissues (P < 0.05, Fig. 1A). Subsequently, we tested circ-0010117 expression in U251 and U87 cells. Synthesized interfering oligonucleotides targeting circ-0010117 significantly decreased the expression of circ-0010117 in U251 and U87 cell lines (Fig. 1B and C). Circ-0010117 overexpression strongly increased the expression of circ-0010117 in U251 and U87 cell lines (Fig. 1D).
Fig. 1.
Circ-0010117 is downregulated in glioblastoma. (A) Circ-0010117 mRNA levels in paratumoural and tumor tissues were determined by qRT‒PCR. *P < 0.05 vs. paratumoural tissues. Circ-0010117 expression in U251 (B) and U87 (C) cells. *P < 0.05 vs. NC. (D) Synthesized interfering oligonucleotides targeting circ-0010117 significantly decreased the expression of circ-0010117 in U251 and U87 cell lines. *P < 0.05 vs. Control.
Circ-0010117 regulates aggressiveness in glioblastoma
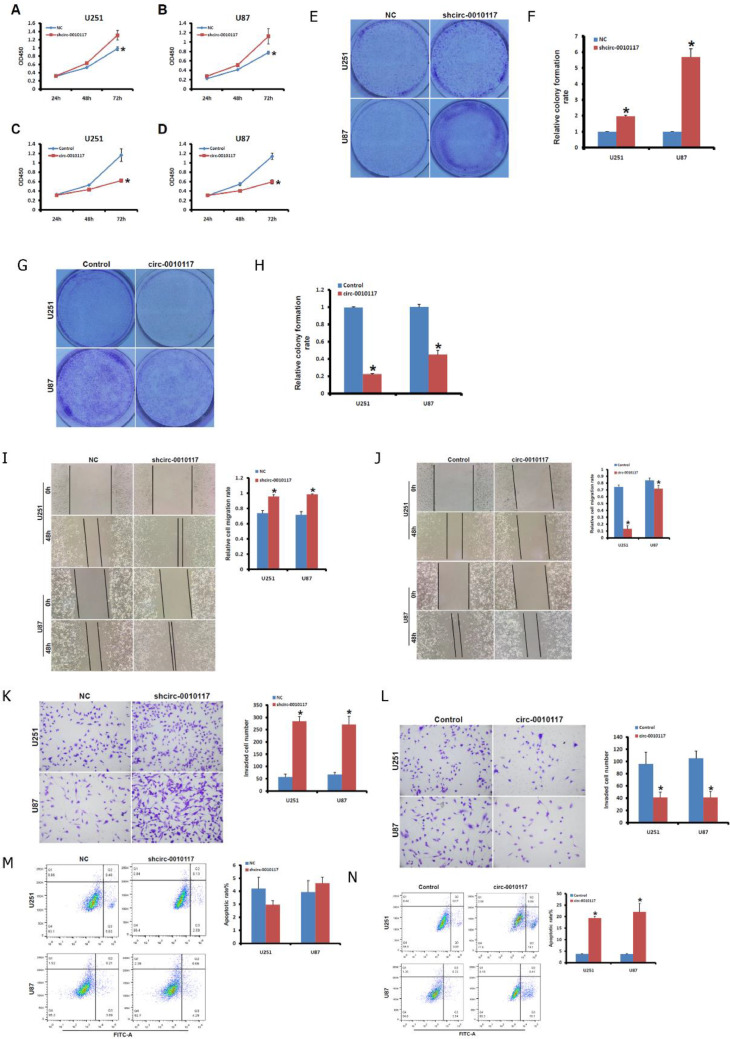
The silencing of circ-0010117 in U251 and U87 cell lines remarkably enhanced cell proliferation compared with the control (Fig. 2A and B). However, the overexpression of circ-0010117 in U251 and U87 cell lines significantly suppressed cell proliferation (Fig. 2C and D). We next sought to establish whether alteration of circ-0010117 could affect U251 and U87 clonogenicity. As shown in Fig. 2E–H, circ-0010117 silencing significantly enhanced the anchorage-dependent growth of MSTO-211H cells (Fig. 2E and F). In addition, circ-0010117 overexpression strongly decreased the anchorage-dependent growth of U251 and U87 cells (Fig. 2G and H). Thus, circ-0010117 can regulate growth activities in glioblastoma cells.
Fig. 2.
Circ-0010117 modulates aggressiveness in glioblastoma. The effect of shcirc-0010117 on the proliferation of U251 (A) and U87 (B) cells was detected with a CCK-8 assay. *P < 0.05 vs. NC. The effect of circ-0010117 overexpression on the proliferation of U251 (C) and U87 (D) cells was detected with a CCK-8 assay. *P < 0.05 vs. NC. The effect of shcirc-0010117 on colony formation of U251 and U87 cells (E, F) was detected by a colony formation assay. *P < 0.05 vs. NC. The effect of circ-0010117 overexpression on colony formation of U251 and U87 cells (G, H) was detected by a colony formation assay. *P < 0.05 vs. NC. The effect of shcirc-0010117 on wound healing of U251 and U87 cells (I). *P < 0.05 vs. NC. The effect of circ-0010117 overexpression on wound healing of U251 and U87 cells (J). *P < 0.05 vs. NC. The effect of shcirc-0010117 on the invasion of U251 and U87 cells (K) was detected by transwell assay. *P < 0.05 vs. NC. The effect of circ-0010117 overexpression on the invasion of U251 and U87 cells (L) was detected by transwell assay. *P < 0.05 vs. NC. The effect of shcirc-0010117 on the apoptosis of U251 and U87 cells (M) was detected by flow cytometry. *P < 0.05 vs. NC. The effect of circ-0010117 overexpression on the apoptosis of U251 and U87 cells (N) was detected by flow cytometry. *P < 0.05 vs. NC.
Enhanced migration and invasion are another key feature across the metastatic cascade. To assess the effect of circ-0010117 on the migration and invasion of MPM cells, wound scratch and transwell assays were performed, respectively. As shown in Fig. 2I, the rate of gap closure was greatly enhanced after circ-0010117 silencing in U251 and U87 cells compared with the control. However, the rate of gap closure was significantly decreased after circ-0010117 overexpression in U251 and U87 cells compared with the control (Fig. 2J).
Similarly, a markedly increased cell invasion was observed in U251 and U87 cells with circ-0010117 silencing compared with the control by transwell assay (Fig. 2K). In contrast, we observed significantly decreased cell invasion in U251 and U87 cells overexpressing circ-0010117 compared with the control (Fig. 2L). We also assessed apoptosis alteration by circ-0010117 regulation through flow cytometry. U251 and U87 cells exhibited a reduced percentage of apoptotic cells after circ-0010117 silencing (Fig. 2M). For U251 and U87 cells overexpressing circ-0010117, the percentage of cells exhibiting apoptosis was significantly increased (Fig. 2N). These results suggest that circ-0010117 regulates aggressiveness in glioblastoma.
Circ-0010117 regulates aggressiveness via miRNA-6779-5p
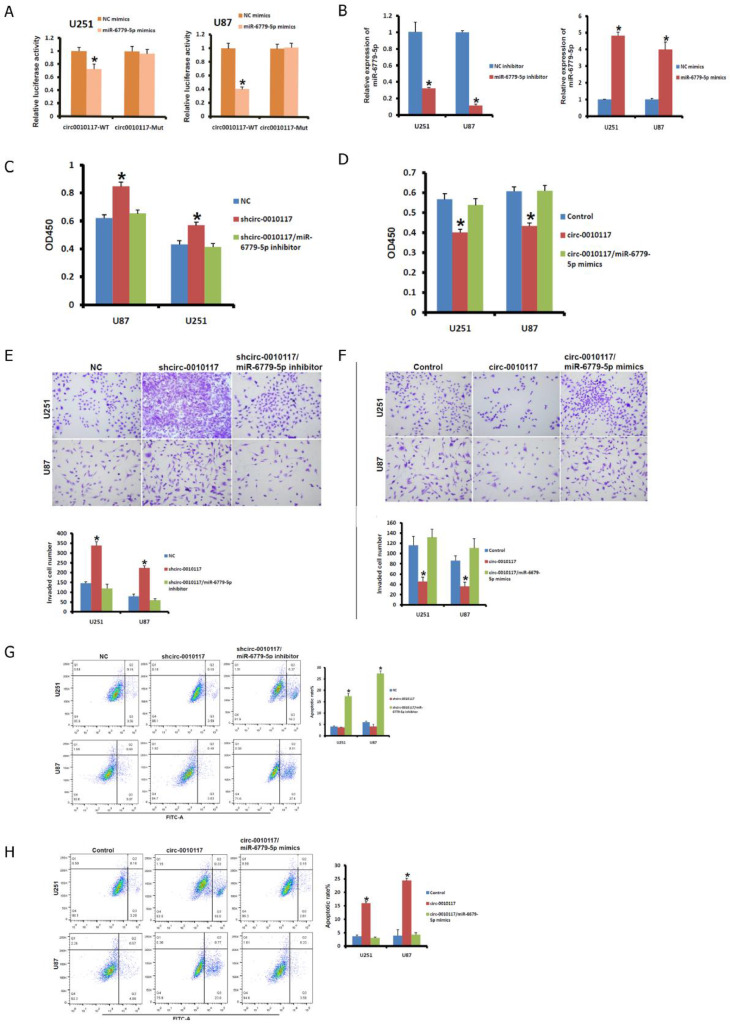
To unravel the relevant mechanism underlying circ-0010117 contributing to the malignant phenotype of glioblastoma. We searched for a potential regulatory mechanism of circ-0010117. We next asked whether miRNAs play important roles in the regulation of circ-0010117 related to the malignant phenotype of glioblastoma. We found that miR-6779-5p significantly reduced the luciferase activities of circ-0010117 WT but not Mut in U251 and U87 cells (Fig. 3A). We next investigated the effect of inhibiting or overexpressing miR-6779-5p in U251 and U87 cells with circ-0010117 silencing or circ-0010117 overexpression. The miR-6779-5p expression level was confirmed after inhibiting or overexpressing miR-677-5p (Fig. 3B).
Fig. 3.
Circ-0010117 regulates aggressiveness via miRNA-6779-5p. MiR-6779-5p significantly reduced the luciferase activities of circ-0010117 WT but not Mut in U251 (A) and U87 (B) cells. *P < 0.05 vs. NC mimics. (B) The miR-6779-5p expression level was confirmed after inhibiting or overexpressing miR-677-5p. *P < 0.05 vs. NC inhibitor or NC mimics. (C) The effect of shcirc-0010117 and miR-6779-5p inhibitor on the proliferation of U251 and U87 cells was detected with CCK-8 assays. *P < 0.05 vs. NC. (D) The effect of circ-0010117 and miR-6779-5p overexpression on the proliferation of U251 and U87 cells was detected with CCK-8 assays. *P < 0.05 vs. NC. (E) The effect of shcirc-0010117 and miR-6779-5p inhibitor on the invasion of U251 and U87 cells was detected with a transwell assay. *P < 0.05 vs. NC. (F) The effect of circ-0010117 and miR-6779-5p overexpression on the invasion of U251 and U87 cells was detected with a transwell assay. *P < 0.05 vs. NC. (G) The effect of shcirc-0010117 and miR-6779-5p inhibitor on U251 and U87 cell apoptosis was detected with flow cytometry. *P < 0.05 vs. NC. (H) The effect of circ-0010117 and miR-6779-5p overexpression on the apoptosis of U251 and U87 cells was detected with flow cytometry. *P < 0.05 vs. NC.
Shcirc-0010117 and miR-6779-5p inhibitor were transfected into U251 and U87 cells to knockdown circ-0010117 and miR-6779-5p expression. The effect of the miR-6779-5p inhibitor on U251 and U87 cells was consistent with the effect of circ-0010117 on cell proliferation detected with CCK-8 (Fig. 3C). In contrast, circ-0010117 and miR-6779-5p mimic were transfected into U251 and U87 cells to overexpress circ-0010117 and miR-6779-5p. We found that miR-6779-5p overexpression enhanced U251 and U87 cell proliferation, which reversed the effect of circ-0010117 overexpression on cell proliferation (Fig. 3D).
Similar results were found for invasion and apoptosis. The effect of the miR-6779-5p inhibitor on U251 and U87 cells was consistent with the effect of circ-0010117 on cell invasion and apoptosis detected with transwell assays and flow cytometry (Fig. 3E and G). In addition, we found that miR-6779-5p overexpression enhanced U251 and U87 cell invasion and apoptosis, which reversed the effect of circ-0010117 overexpression on cell invasion and apoptosis in cells overexpressing circ-0010117 and miR-6779-5p (Fig. 3F and H).
SPEN contributed to miRNA-6779-5p regulation
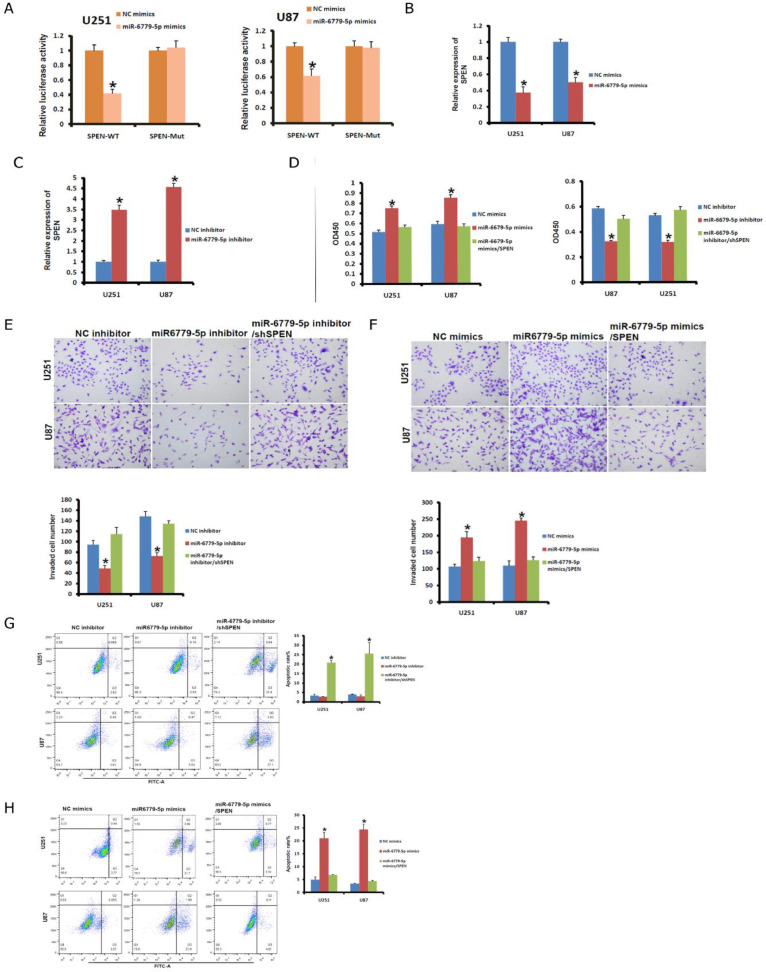
To explore the regulatory mechanism underlying circ-0010117 and miR-6779-5p for the malignant phenotype of glioblastoma. We subsequently identified a potential target of miR-6779-5p. We found that miR-6779-5p significantly reduced the luciferase activities of SPEN WT but not Mut in U251 and U87 cells (Fig. 4A). The SPEN mRNA level was also found to be significantly decreased after miR-6779-5p inhibition in U251 and U87 cells (Fig. 4B) but markedly increased after miR-6779-5p overexpression (Fig. 4C).
Fig. 4.
SPEN contributed to miRNA-6779-5p regulation. MiR-6779-5p significantly reduced the luciferase activities of SPEN WT but not Mut in U251 (A) and U87 (B) cells. *P < 0.05 vs. NC mimics. (B) The SPEN expression level was confirmed after overexpressing (C) or inhibiting (D) miR-677-5p. *P < 0.05 vs. NC mimics or NC inhibitor. (D) The effects of miR-6779-5p and SPEN overexpression on the proliferation of U251 and U87 cells were detected with CCK-8 assays (left). The effect of the miR-6779-5p inhibitor and shSPEN on the proliferation of U251 and U87 cells was detected with CCK-8 assays (right). *P < 0.05 vs. NC mimic or NC inhibitor. (E) The effect of the miR-6779-5p inhibitor and shSPEN on the invasion of U251 and U87 cells was detected with a transwell assay. *P < 0.05 vs. NC inhibitor. (F) The effect of miR-6779-5p and SPEN overexpression on the invasion of U251 and U87 cells was detected with a transwell assay. *P < 0.05 vs. NC mimic. (G) The effect of the miR-6779-5p inhibitor and shSPEN on the apoptosis of U251 and U87 cells was detected with flow cytometry. *P < 0.05 vs. NC inhibitor. (H) The effect of miR-6779-5p and SPEN overexpression on the apoptosis of U251 and U87 cells was detected with flow cytometry. *P < 0.05 vs. NC mimic.
The miR-6779-5p mimic and SPEN overexpression plasmid were transfected into U251 and U87 cells to increase miR-6779-5p and SPEN expression. SPEN overexpression in U251 and U87 cells reversed the effect of miR-6779-5p overexpression on cell proliferation detected with CCK-8 (Fig. 4D). We also transfected U251 and U87 cells with miR-6779-5p inhibitor and shSPEN to knock down miR-6779-5p and SPEN. As shown in Fig. 4D, SPEN knockdown blocked the effect of miR-6779-5p inhibition on cell proliferation suppression.
Consistent with cell proliferation, SPEN knockdown reversed the effect of miR-6997-5p silencing on invasion and apoptosis in U251 and U87 cells (Fig. 4E and G). However, SPEN overexpression repressed U251 and U87 cell invasion and apoptosis caused by miR-6779-5p overexpression (Fig. 4F and H).
Circ-0010117 upregulation inhibited tumor growth in vivo
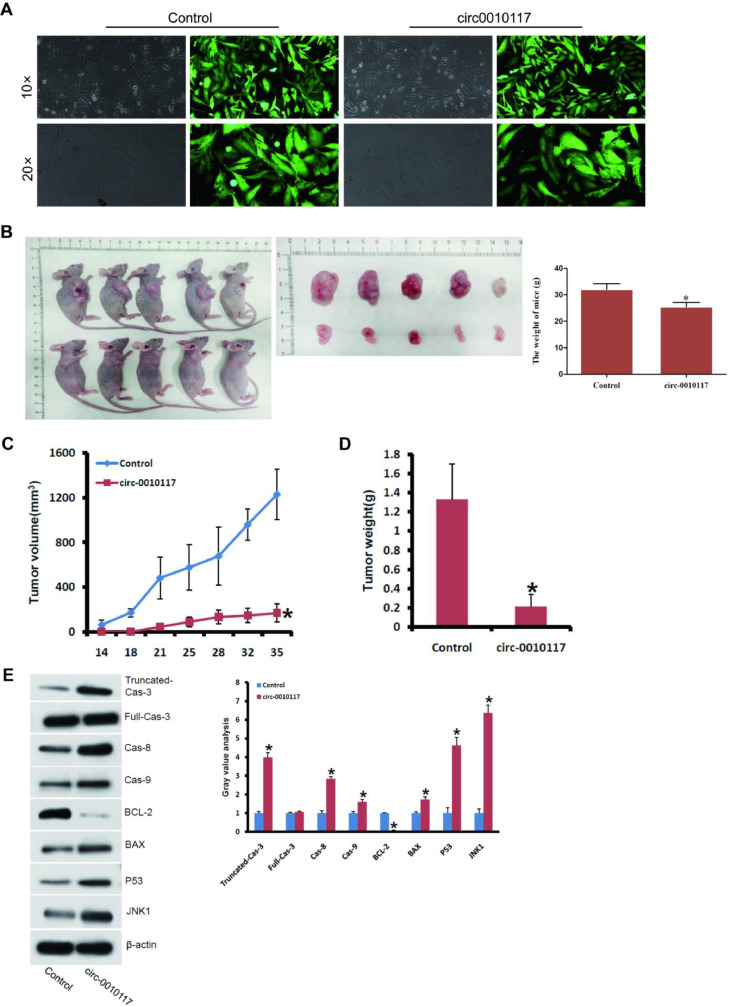
To further assess whether overexpression of circ-0010117 could suppress tumor formation in vivo, we inoculated stable overexpressed circ-0010117 with the U251 cell line (Fig. 5A) subcutaneously into 4-week-old nude mice. The results showed that tumor size and weight were remarkably reduced when transplanted with tumor cells overexpressing U251 (Fig. 5B–D).
Fig. 5.
Circ-0010117 upregulation inhibited tumor growth in vivo. (A) circ-0010117 is stably overexpressed in the U251 cell line. (B) Subcutaneously injected into 4-week-old nude mice. The results showed that tumor size (B, C) and weight (D) were remarkably reduced when transplanted with tumor cells overexpressing circ-0010117. *P < 0.05 vs. control. (E) The influence of circ-0010117 overexpression on the caspase pathway was determined by western blotting. *P < 0.05 vs. control.
To investigate the molecular mechanism by which circ-0010117 overexpression influences the caspase pathway, we determined each component involved in the pathway by western blotting (Fig. 5E). Truncated-cas-3, full-cas-3, cas-8, cas-9, BAX, P53, and JNK1 were enhanced by circ-0010117 overexpression (Fig. 5E). However, BCL-2 was suppressed by circ-0010117 overexpression (Fig. 5E). Therefore, we concluded that overexpression of circ-0010117 also suppressed tumorigenesis through the caspase pathway in vivo.
Discussion
Currently, glioblastoma is a common malignant tumor worldwide [13]. In recent years, with research on biomarkers of glioblastoma, the targeted diagnosis, treatment and prognosis of glioblastoma have achieved preliminary results, but more research is needed to combat glioblastoma. Therefore, the identification of key biomarkers in glioblastoma is an effective strategy for the treatment of glioblastoma.
The roles of circRNAs in cancer progression have attracted much attention due to their key roles in human cancers. In this study, our goal was to explore the potential important roles of circ-0010117 during glioblastoma progression. This examination confirmed that circ-0010117 participated in the glioblastoma process via miRNA-6779-5p and SPEN, revealing that circ-0010117 served as a novo biomarker for glioblastoma.
CircRNAs represent a new class of endogenous noncoding RNAs that have recently been recognized as important regulators of glioblastoma. MiRNAs are a group of 19–22 nucleotide noncoding, single-stranded RNA molecules that participate in sequence-specific posttranscriptional regulation of gene expression [14]. Recently, emerging evidence has confirmed that circRNAs can regulate the expression of tumor-suppressive or oncogenic genes through the circRNA-miRNA‒mRNA axis. For instance, circular RNA circ-0001946 has been validated to act as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1 [15]. Circular RNA circMTO1 inhibits the proliferation of glioblastoma cells via the miR-92/WWOX signaling pathway [16]. Because their expression and function in glioblastoma development are still largely elusive, we screened the differentially expressed circRNAs between glioblastoma and matched nontumor liver tissues and focused on the role and underlying mechanism of the decreased circ-0010117 in glioblastoma progression. In this study, we found that circ-0010117 is downregulated in glioblastoma. In addition, circ-0010117 can regulate aggressiveness in glioblastoma. This invention expanded knowledge on the role of circ-0010117 in the modulation of glioblastoma, which may be helpful to better elucidate the pathological mechanism of glioblastoma.
Many researchers have illustrated that circRNAs regulate the process of diseases via miRNAs [17]. Previous researchers found that many miRNAs were involved in glioblastoma; for instance, miRNA-381 inhibited EMT in glioblastoma [18]. miRNA-124-3p was reported to suppress the growth of glioblastoma [19]. In contrast, miRNA-1908 promoted glioblastoma [20]. Here, experimental proof identified miRNA-6779-5p as downstream of circ-0010117, as they have confirmed connection points. We found that the level of miRNA-6779-5p was clearly upregulated and was in contrast with the level of circ-0010117 in glioblastoma.
Split ends (SPEN) is an ERα corepressor that we have identified as a tumor suppressor protein in ERα-positive breast cancer cells [21]. SPEN has been reported to play important roles in cancer progression. For instance, SPEN has been found to induce miR-4652-3p to target HIPK2 in nasopharyngeal carcinoma [22]. SPEN has also been identified as a crucial factor for Xist function through forward genetic screening in haploid embryonic stem cells [23]. However, it is still unclear whether SPEN contributes to the circ-0010117 and miR-6779-5p regulatory network. In this study, we found that SPEN is a direct target of miR-6779-5p and that SPEN contributes to the miRNA-6779-5p regulatory network.
Conclusion
In summary, we determined that circ-0010117 is linked to aggressiveness in glioblastoma through miRNA-6779-5p and SPEN. Our results provide the rationale for the use of circ-0001946 as a novel potential therapeutic target in glioblastoma.
Data availability
All the data from the current study are included in the article or uploaded as supplementary information.
Funding
The study was supported by Science and technology project of Health Commission of Jiangxi Province (No: 202210394).
Ethics approval and consent to participate
The experimental protocols were approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University. This paper has not been published elsewhere in whole or in part. All authors have read and approved the content and agree to submit it for consideration for publication in your journal. There are no ethical/legal conflicts involved in the article. Informed consent was obtained from all individual participants included in the study.
CRediT authorship contribution statement
Xuanyong Yang: Writing – original draft, Writing – review & editing, Methodology. Yue Liu: Writing – original draft, Writing – review & editing. Xinhui Zhou: Data curation, Visualization. Kang Chen: Conceptualization, Validation, Supervision. Jiang Xu: Visualization, Data curation. Shan Xu: Writing – original draft, Writing – review & editing, Methodology, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no conflicts of interest.
Acknowledgment
Not applicable.
Contributor Information
Jiang Xu, Email: xujiang81@163.com.
Shan Xu, Email: xshan2002@126.com.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Wang M., Yu F., Wu W., Zhang Y., Chang W., Ponnusamy M., Wang K., Li P. Circular RNAs: a novel type of non-coding RNA and their potential implications in antiviral immunity. Int. J. Biol. Sci. 2017;13(12):1497–1506. doi: 10.7150/ijbs.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao D., Hu S., Zheng X., Lin W., Gao J., Chang K., Zhao D., Wang X., Zhou J., Lu S., Griffiths H.R., Liu J. SOD3 is secreted by adipocytes and mitigates high-fat diet-induced obesity, inflammation, and insulin resistance. Antioxid. Redox Signal. 2020;32(3):193–212. doi: 10.1089/ars.2018.7628. [DOI] [PubMed] [Google Scholar]
- 4.Zheng H., Chen T., Li C., Xu C., Ding C., Chen J., Ju S., Zhang Z., Liang Z., Cui Z., Zhao J. A circular RNA hsa_circ_0079929 inhibits tumor growth in hepatocellular carcinoma. Cancer Manag. Res. 2019;11:443–454. doi: 10.2147/CMAR.S189338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu F., Cheng C., Qin H., Wang H., Yu H. A novel circular RNA circENTPD7 contributes to glioblastoma progression by targeting ROS1. Cancer Cell Int. 2020;20:118. doi: 10.1186/s12935-020-01208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia W., Qiu M., Chen R., Wang S., Leng X., Wang J., Xu Y., Hu J., Dong G., Xu P.L., Yin R. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci. Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 8.Franken N.A., Rodermond H.M., Stap J., Haveman J., van Bree C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006;1(5):2315–2319. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 9.Arsic N., Bendris N., Peter M., Begon-Pescia C., Rebouissou C., Gadea G., Bouquier N., Bibeau F., Lemmers B., Blanchard J.M. A novel function for cyclin A2: control of cell invasion via RhoA signaling. J. Cell Biol. 2012;196(1):147–162. doi: 10.1083/jcb.201102085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L.M., Wang W., Lee J.C., Chiu F.H., Wu C.T., Tai C.J., Wang C.K., Tai C.J., Huang M.T., Chang Y.J. Thrombomodulin mediates the progression of epithelial ovarian cancer cells. Tumour Biol. 2013;34(6):3743–3751. doi: 10.1007/s13277-013-0958-x. [DOI] [PubMed] [Google Scholar]
- 11.Espada J., Matabuena M., Salazar N., Lucena S., Kourani O., Carrasco E., Calvo M., Rodriguez C., Reyes E., Gonzalez S., Juarranz A. Cryptomphalus aspersa mollusc eggs extract promotes migration and prevents cutaneous ageing in keratinocytes and dermal fibroblasts in vitro. Int. J. Cosmet. Sci. 2015;37(1):41–55. doi: 10.1111/ics.12167. [DOI] [PubMed] [Google Scholar]
- 12.Lee S.J., Kim W.J., Moon S.K. Role of the p38 MAPK signaling pathway in mediating interleukin-28A-induced migration of UMUC-3 cells. Int. J. Mol. Med. 2012;30(4):945–952. doi: 10.3892/ijmm.2012.1064. [DOI] [PubMed] [Google Scholar]
- 13.Omuro A. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 14.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 15.Li X., Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR-671-5p and CDR1. J. Cell. Physiol. 2019;234(8):13807–13819. doi: 10.1002/jcp.28061. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X., Zhong B., Zhang W., Wu J., Wang Y. Circular RNA CircMTO1 inhibits proliferation of glioblastoma cells via miR-92/WWOX signaling pathway. Med. Sci. Monit. 2019;25:6454–6461. doi: 10.12659/MSM.918676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang X. miRNA, lncRNA and circRNA: targeted molecules full of therapeutic prospects in the development of diabetic retinopathy. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.771552. (Lausanne) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min RQ. MicroRNA-381 inhibits metastasis and epithelial-mesenchymal transition of glioblastoma cells through targeting LEF1. Eur. Rev. Med. Pharmacol. Sci. 2020;24(12):6825–6833. doi: 10.26355/eurrev_202006_21672. [DOI] [PubMed] [Google Scholar]
- 19.S Cai. miR-124-3p inhibits the viability and motility of glioblastoma multiforme by targeting RhoG. Int. J. Mol. Med. 2021;47(5):69. doi: 10.3892/ijmm.2021. [DOI] [PubMed] [Google Scholar]
- 20.X Xia. MicroRNA-1908 functions as a glioblastoma oncogene by suppressing PTEN tumor suppressor pathway. Mol. Cancer. 2015;14:154. doi: 10.1186/s12943-015-0423-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legare S., Chabot C., Basik M. SPEN, a new player in primary cilia formation and cell migration in breast cancer. Breast Cancer Res. 2017;19(1):104. doi: 10.1186/s13058-017-0897-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., Lv Y., Cheng C., Huang Y., Yang L., He J., Tao X., Hu Y., Ma Y., Su Y., Wu L., Yu G., Jiang Q., Liu S., Liu X., Liu Z. SPEN induces miR-4652-3p to target HIPK2 in nasopharyngeal carcinoma. Cell Death Dis. 2020;11(7):509. doi: 10.1038/s41419-020-2699-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monfort A., Di Minin G., Postlmayr A., Freimann R., Arieti F., Thore S., Wutz A. Identification of SPEN as a crucial factor for xist function through forward genetic screening in haploid embryonic stem cells. Cell Rep. 2015;12(4):554–561. doi: 10.1016/j.celrep.2015.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data from the current study are included in the article or uploaded as supplementary information.