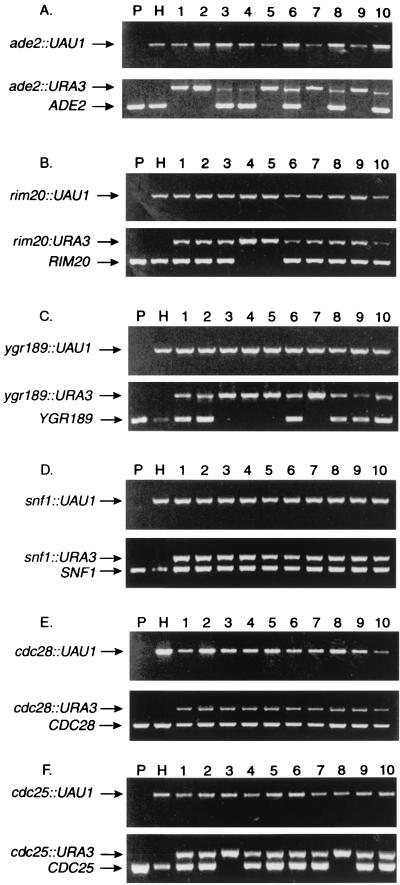
FIG. 2.
PCR analysis of double-disruption selection segregants. Each panel shows a composite of two agarose gels of PCRs conducted with genomic DNA templates. The upper-gel PCRs used amp3 and Arg4det primers; the lower-gel PCRs used amp3 and amp5 primers. (The amp3–amp5 PCRs are unreliable for detection of full-length UAU1 insertions, presumably because the smaller PCR products are amplified more efficiently.) Lanes: templates from parent strain BWP17 (lanes P), the respective UAU1 disruption heterozygote (lanes H), and 10 independent Arg+ Ura+ segregants from the disruption heterozygote (lanes 1 to 10). Panels show analyses for ADE2 (A), RIM20 (B), YGR189 (C), SNF1 (D), CDC28 (E), and CDC25 (F).