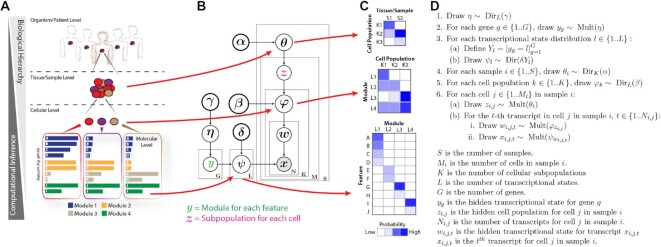
Figure 1.
Celda identifies cell heterogeneity by clustering genes into modules and cells into subpopulations. (A) Example of a biological hierarchy. One way in which we try to understand complex biological systems is by organizing them into hierarchies. Individual organisms are composed of complex tissues. Each complex tissue is composed of different cellular populations with distinct functions; each cellular subpopulation contains a unique mixture of molecular pathways (i.e. modules); and each module is composed of groups of genes that are co-expressed across cells. (B) Plate diagram of the Celda_CG model. We developed a novel discrete Bayesian hierarchical model called Celda_CG to characterize the molecular and cellular hierarchies in biological systems. Celda_CG performs ‘co-clustering’ by assigning each gene to a module and each cell to a subpopulation. (C) In addition to clustering, Celda_CG also inherently performs a form of ‘matrix factorization’ by deriving three distinct probability matrices: (i) a cell population × sample matrix representing the probability that each population is present in each sample; (ii) a transcriptional module × cell population matrix representing the contribution of each transcriptional state to each cellular subpopulation; and (iii) a gene × module matrix representing the contribution of each gene to its module. (D) Generative process for the Celda_CG model.