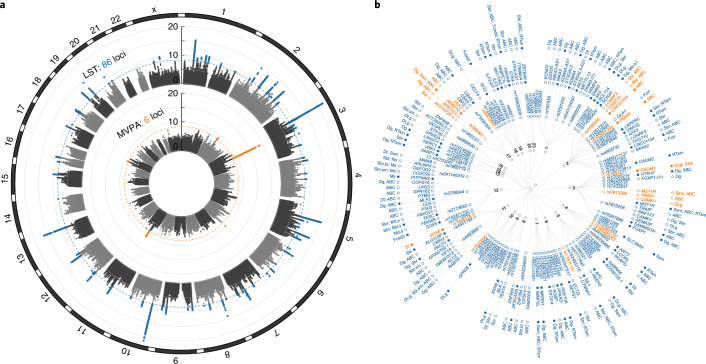
Fig. 2. Main results of GWAS and downstream gene prioritization for LST and MVPA.
a, Circular Manhattan plot summarizing the results from European ancestry meta-analyses for LST and MVPA. Outer track, LST; inner track, MVPA. Genome-wide significant variants (P < 5 × 10−9) are highlighted in orange for loci associated with MVPA and in blue for loci associated with LST. b, Dendrogram showing the 101 independent association signals in LST- and MVPA-associated loci from European ancestry or multi-ancestry meta-analyses. Moving outwards from the center are: (1) chromosome; (2) lead SNP identifiers, in orange for loci associated with MVPA, in blue for loci associated with LST; (3) the most promising gene(s) prioritized in the locus (closest genes are highlighted by filled circles); and (4) the approach(es) by which the gene was prioritized, that is, DEPICT gene prioritization (Dg) or tissue enrichment (Dt); SMR of eQTL signals in blood (Sbl), brain (Sbr) or skeletal muscle (Ssm); credible variants identified by FINEMAP that (i) are coding and likely to have a detrimental effect on protein function (Fcadd) or (ii) show evidence of three-dimensional interactions with the candidate gene in central nervous system cell types (Fcrt); activity-by-contact (ABC) in 26 relevant tissues and cell types; a contribution to enrichment for altered expression in skeletal muscle following a resistance training intervention (RTsm); and/or proximity to an association signal for spontaneous running speed (Ms), time run (Mt) or distance run (Md) in a GWAS of 100 inbred mouse strains.