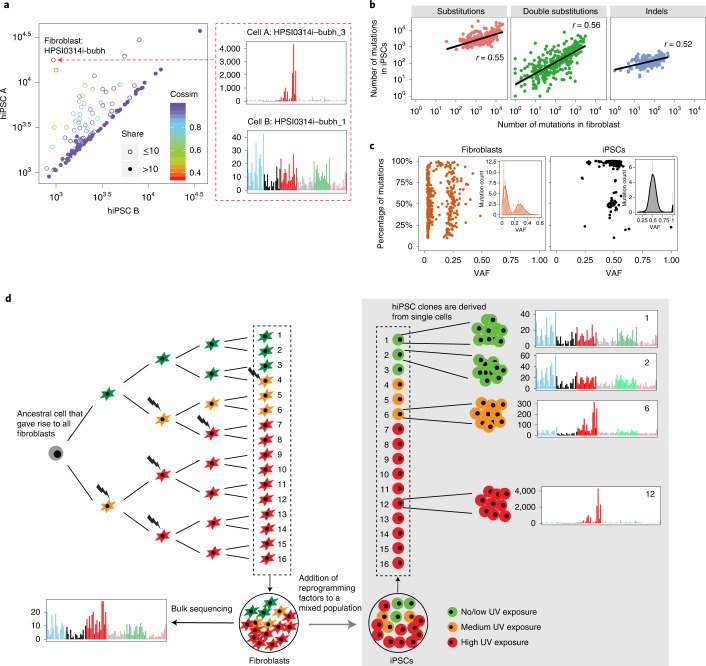
Fig. 3. Genomic heterogeneity in F-hiPSCs.
a, Comparison of substitutions carried by pairs of hiPSCs that were derived from fibroblasts of the same donor and in the same reprogramming experiment. Each circle represents a single donor. Hollow circles indicate that two hiPSCs from the same donor share fewer than ten mutations, whereas filled circles indicate that they share ten or more mutations. Colors denote cosine similarity values between mutation profiles of two hiPSCs. A very high score (purple hues) indicates a strong likeness. n = 164 fibroblasts with two derived hiPSCs. b, Correlations between number of mutations in hiPSCs and their matched fibroblasts, n = 324 WGS of hiPSCs. c, Summary of subclonal clusters in fibroblasts (n = 204 WGS of fibroblasts) and hiPSCs (n = 324 WGS of hiPSCs). Kernel density estimation was used to smooth the distribution. Local maximums and minimums were calculated to identify subclonal clusters. Each dot represents a cluster that has at least 10% of total mutations in the sample. Most fibroblasts are polyclonal with a cluster VAF near 0.25, whereas hiPSCs are mostly clonal, with VAF near 0.5. d, Schematic illustration of genomic heterogeneity in F-hiPSCs. Through bulk sequencing of fibroblasts, all of these cells will carry all the mutations that were present in the gray cell, their most recent common ancestor. The individual cells will also carry their unique mutations depending on the DNA damage received by each cell. Each hiPSC clone is derived from a single cell. Subclone 1 and subclone 2 cells are more closely related and could share a lot of mutations in common, because they share a more recent common ancestor. However, they are distinct cells and will create separate hiPSC clones. Subclone 6 (orange cells) and subclone 12 (red cells) are not closely related to the green cells and have received more DNA damage from UV, making them genomically divergent from the green cells. They could still share some mutations in common, because they shared a common ancestor at some early point, but will have many of their own unique mutations (largely due to UV damage).