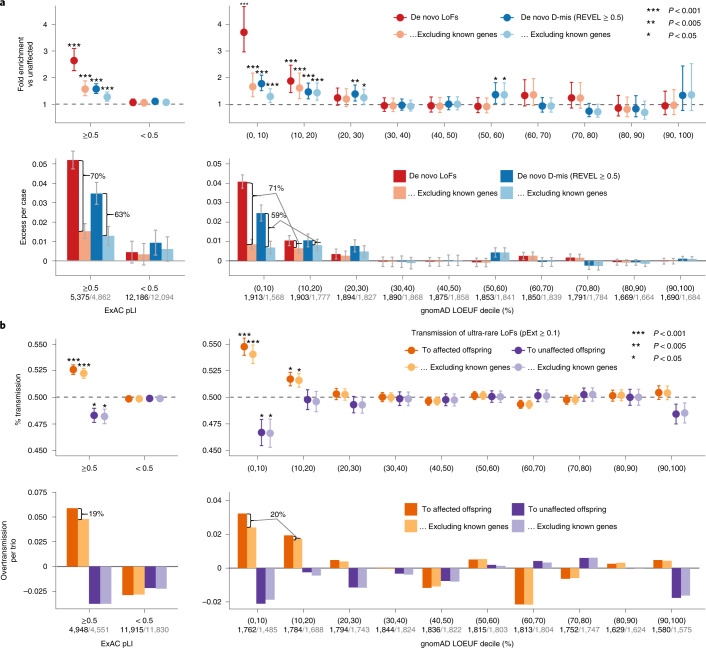
Fig. 2. Comparison of burden between de novo damaging variants and rare inherited LoFs in ASD.
a, The burden of DNVs was evaluated by the rate ratio and rate difference between 16,877 ASD and 5,764 unaffected trios. The exome-wide burden of de novo LoF and D-mis (REVEL ≥ 0.5) variants are concentrated in constrained genes (ExAC pLI ≥ 0.5) and in genes with the highest levels of LoF intolerance in the population (defined by the top two deciles of gnomAD LOEUF scores). Burden analysis was repeated after removing known ASD or NDD genes. The number of genes before and after removing known genes in each constraint bin is shown below the axis label. Data are presented as mean values and 95% confidence intervals. Among constrained genes (ExAC pLI ≥ 0.5 or the top 20% of gnomAD LOEUF scores), close to two-thirds of case-control rate differences of de novo LoF and D-mis variants can be explained by known genes. Exact P values by Poisson test are listed in Supplementary Table 19. b, The burden of inherited LoFs was evaluated by looking at the proportion of rare LoFs in 20,491 parents without ASD diagnoses or intellectual disability that are transmitted to affected offspring in 9,504 trios and 2,966 duos and show evidence of overtransmission of LoFs per ASD trio. As a comparison, we also show the transmission disequilibrium pattern to unaffected offspring in 5,110 trios and 129 duos. Data are presented as mean values ± standard errors as error bars. Two-sided binomial test was used to compute the P values for overtransmission or undertransmission. Using ultra-rare LoFs with pExt ≥ 0.1, exome-wide signals of transmission disequilibrium of rare inherited LoF variants also concentrate in constrained genes (ExAC pLI ≥ 0.5) and in genes within the top two deciles of gnomAD LOEUF scores. Analysis was restricted to autosomal genes and repeated after removing known ASD or NDD genes (number of genes in each constrained bin before and after removing known genes is shown below the axis label). Among all constrained genes, only one-fifth of overtransmission of LoFs to ASD trios can be explained by known ASD or NDD genes. Exact P values by binominal test are listed in Supplementary Table 19.