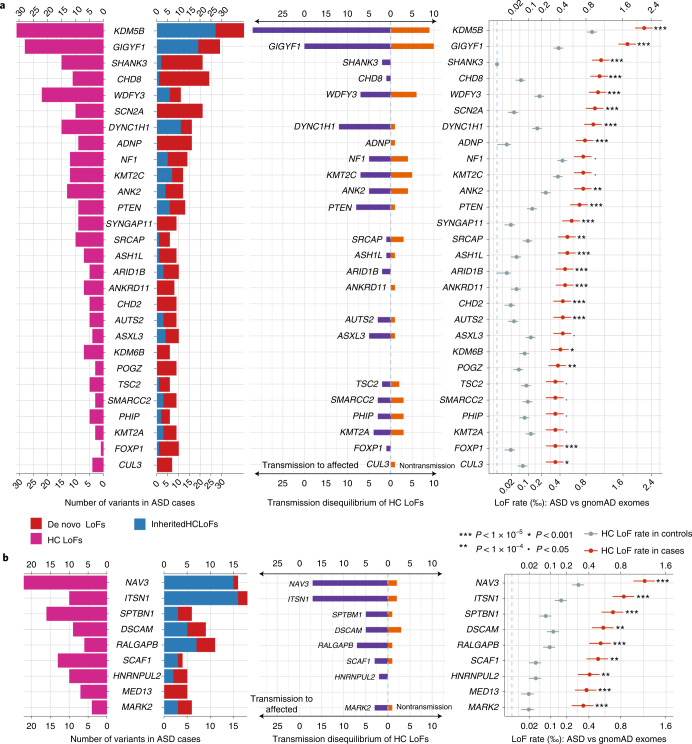
Fig. 5. Distribution of de novo and inherited LoF variants in known and novel ASD genes in cases and population controls.
From left to right: pyramid plots summarizing the number of de novo LoFs in 15,857 ASD trios, inherited high-confidence LoFs in 18,720 unrelated offspring included in transmission analysis, and high-confidence LoFs in 15,780 unrelated cases; bar plot of transmission vs nontransmission for rare high-confidence LoFs identified in parents without ASD diagnoses or intellectual disability; three plots comparing the high-confidence LoF rate in 31,976 unrelated ASD cases with gnomAD exomes (non-neuro subset, 104,068 individuals). Horizontal bars are presented as mean values ± standard errors as error bars. a, Twenty-eight known ASD or NDD genes that have LOEUF scores in the top 30% of gnomAD, have a P value for enrichment among all DNVs (P < 9 × 10−6) in 23,039 ASD trios, and have more than 10 LoFs. b, Nine additional ASD risk genes that achieved a P value of <9 × 10−6 in stage 2 of this analysis. The majority of genes in b harbor more inherited LoFs than DNVs. All five novel genes (Table 1) are shown in b. Note that the x axes of LoF rates are in the squared root scale. Poisson test was used to compute the P values. Exact P values are listed in Supplementary Table 6.