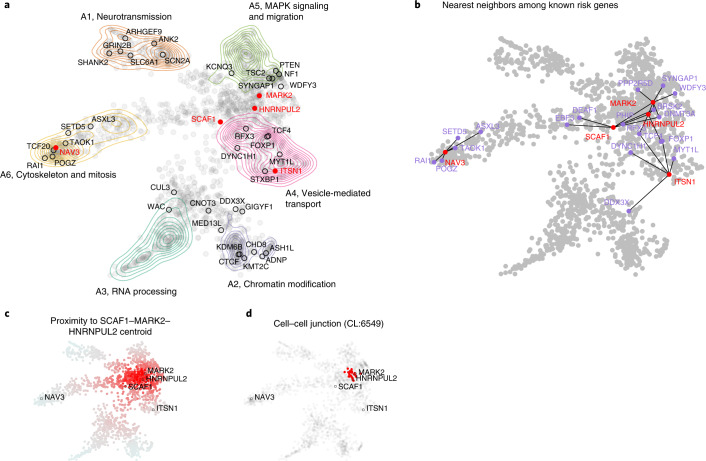
Fig. 7. Functional or phenotypic embedding of ASD risk genes.
a, Using a combination of archetypal analysis and canonical correlation analysis, putative autism risk genes were organized into k = 6 archetypes that represent distinct mechanistic (STRING) and phenotypic (HPO) categorizations (neurotransmission, chromatin modification, RNA processing, vesicle-mediated transport, MAPK signaling and migration, and cytoskeleton and mitosis). Genes implicated by our meta-analysis are indicated by their label, with novel genes in red. b, For each of the five novel genes, we identified the five nearest neighbors in the embedding space among the 62 meta-analysis genes. SCAF1, MARK2 and HNRNPUL2 were identified as ‘mixed’ rather than ‘archetypal’ in their probable risk mechanisms. c, To gain further insight into possible risk mechanisms, we calculated the embedding distance to the centroid of these three genes, which was then used as an index variable to perform gene set enrichment analysis. d, A STRING cluster (CL:6549) containing genes related to cell–cell junctions and the gap junction was identified as being highly localized in this region of the embedding space (P = 4.12 × 10−14 by the Kolmogorov–Smirnov test). This may suggest that these genes confer autism risk through dysregulation of processes related to cell adhesion and migration.