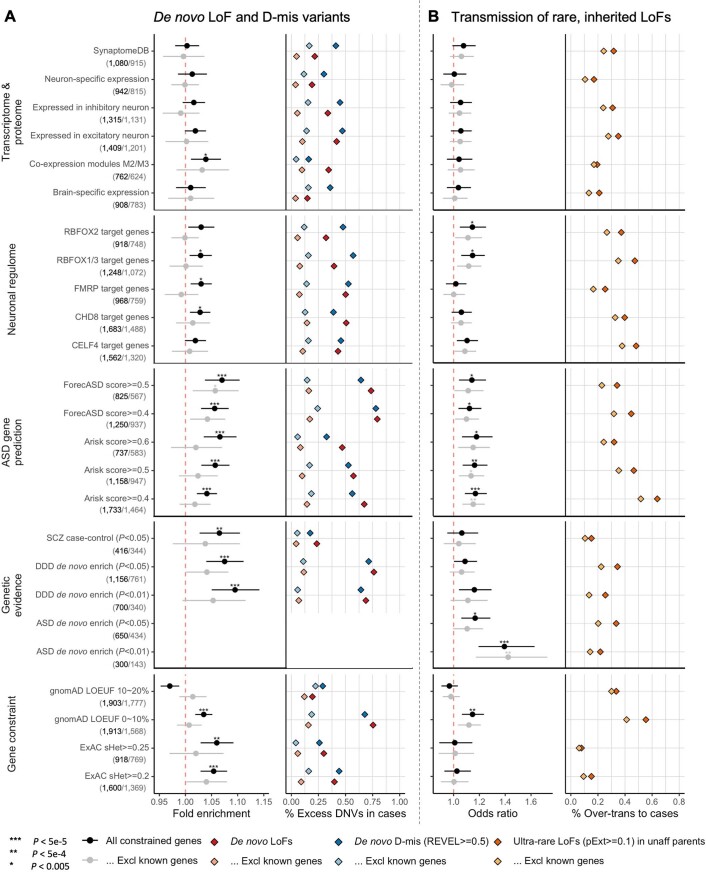
Extended Data Fig. 2. Enrichment of de novo damaging and rare, inherited LoF variants in ASD cases across gene sets.
Gene sets were defined and grouped by transcriptome proteome, neuronal regulome, ASD gene prediction scores, genetic evidence from neuropsychiatric diseases, and gene level constraint. Analyses were repeated after removing known ASD/NDD genes. Number of genes in each set before and after removing known genes are shown in bracket below gene set. Dots represent fold enrichment of DNVs or odds ratio for over-transmission of LoFs in each set. Horizontal bars indicate 95% confidence interval. For each gene set, also shown are the percentage of excessive DNVs in cases and percentage of over-transmission of rare, inherited LoFs to cases. (A) De novo enrichment analysis was performed by dnEnrich that conditional on the overall increase in burden of de novo damaging variants in cases compared with controls (Methods). P-values were derived from 100,000 random permutations of de novo damaging variants among all 5,754 constrained genes and accounts for the tri-nucleotide sequence context and gene length. (B) Enrichment of rare, inherited LoFs was evaluated by comparing the transmission and non-transmission of ultra-rare LoFs with pExt > =0.1 in the gene set versus those in all other constrained genes using a 2-by-2 table. P-values were given by chi-squared test.