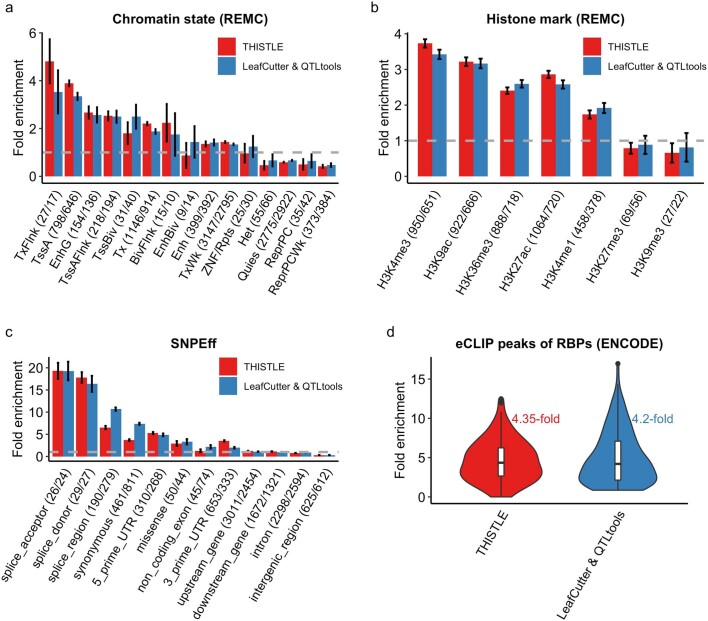
Extended Data Fig. 8. Enrichment of the lead THISTLE or LeafCutter & QTLtools cis-sQTL SNPs in functional annotation categories.
The annotation categories were defined by the chromatin state annotation data from REMC (a), histone marks from REMC (b), predicted variant functions by SNPEff (c), or eCLIP peaks of 113 RBPs binding sites from ENCODE (d). The fold enrichment was computed by dividing the percentage of lead cis-sQTL SNPs in a category by the mean percentage observed in 1,000 sets of control SNPs sampled repeatedly at random (Methods). Each column represents a point estimate with an error bar indicating the 95% CI of the estimate. The grey dashed line represents no enrichment. The numbers in the parentheses are the numbers of THISTLE/LeafCutter & QTLtools sQTL SNPs in each functional category. In panel d, a violin plot shows the distribution of fold enrichment estimates across 113 RBPs binding sites. The line inside each box indicates the median value, notches indicate the 95% CI, the central box indicates the IQR, whiskers indicate data up to 1.5 times the IQR, and outliers are shown as separate dots.