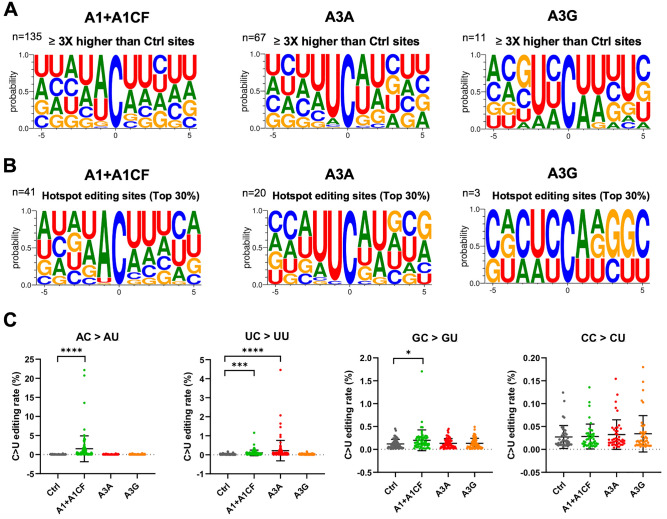
Figure 2.
Local sequence context at the APOBEC-edited C sites on SARS-CoV-2 RNA. (A) Local sequences around the significantly edited target C sites (± 5 nucleotides from target C at position 0) by A1 + A1CF, A3A, or A3G. The editing level of each C site was normalized to the Ctrl, and only sites with 3 × or higher editing levels than the normalized value were defined as significant editing sites. (B) Analysis of local sequences around the top 30% edited C sites (or hotspot editing sites), showing predominantly AC motif for A1 + A1CF, UC for A3A, and CC for A3G. (C) Comparison of the C-to-U editing rates (%) of a particular dinucleotide motif by the three APOBECs. Each dot represents the C-to-U editing level obtained from the SSS results. In panel-D, statistical significance was calculated by unpaired two-tailed student’s t-test with P-values represented as: P > 0.05 = not significant; not indicated, *P < 0.05, ***P < 0.001, ****P < 0.0001.