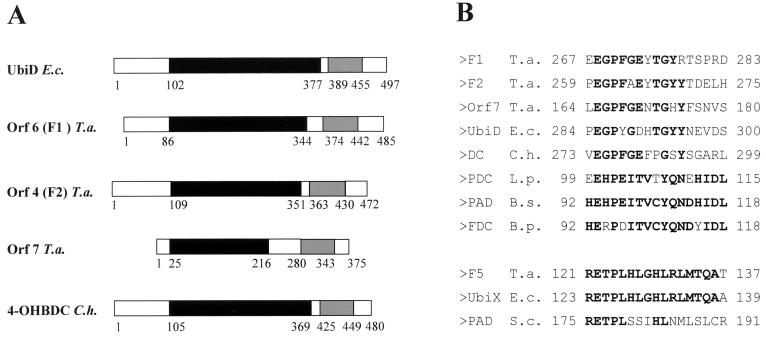
FIG. 8.
Alignment of the conserved regions of F1, F2, and Orf7 possibly involved in phenol metabolism showing similarity to 3-octaprenyl-4-hydroxybenzoate carboxy lyase of E. coli and to 4-hydroxybenzoate decarboxylase of C. hydroxybenzoicum (A) and regions of conserved amino acid sequences in several decarboxylases (B). (A) Boxes with like colors (black and grey) represent similar domains. UbiD, 3-octaprenyl-4-hydroxybenzoate carboxy lyase; 4-OHBDC, 4-hydroxybenzoate decarboxylase; F1, F2, and Orf7, phenol-induced proteins of T. aromatica. (B) PDC, p-coumaric acid decarboxylase; PAD, phenylacrylic acid decarboxylase; FDC, ferulate decarboxylase. Bold letters represent amino acids that are identical in more than 50% of the sequences. The numbers refer to the first and last positions of the motif in the corresponding sequence. E.c., E. coli; T.a., T. aromatica; C.h., C. hydroxybenzoicum; S.c., Saccharomyces cerevisiae; L.p., Lactobacillus plantarum; B.s., Bacillus subtilis; B.p., Bacillus pumilus.