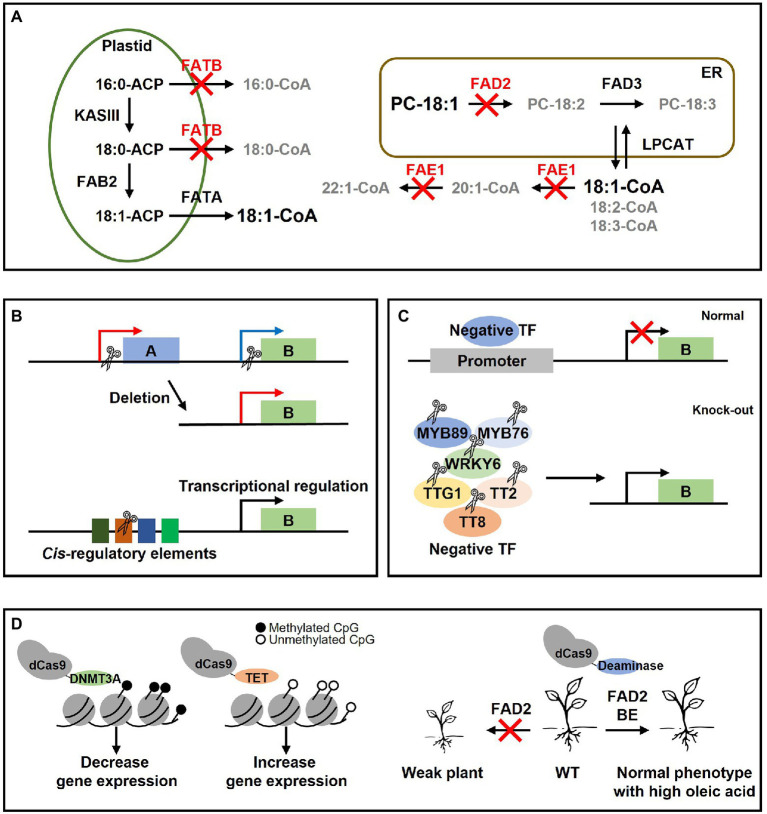
Figure 3.
Future strategies of lipid metabolism research using CRISPR/Cas9 in this paper. (A) Schematic diagram of fatty acid synthesis for the development of plants with high oleic acid content in seeds. The flow of fatty acid synthesis is indicated by arrows. Red letters indicate three genes that may increase the oleic acid content if it is eliminated by CRISPR. Gray letters indicate fatty acids whose content is decreased when three genes (FATB, FAE1, and FAD2) are knocked out. It appears that plants with high oleic acid content may be created if three genes were deleted. (B) A study of promoter regulation using CRISPR/Cas9. B refers to the lipid gene and A is the upstream gene of the B gene. The scissor shape represents CRISPR/Cas9. By removing from the 5′UTR of the B gene to the 5′UTR of the A gene, the B gene can be controlled by the A promoter (Bhunia et al., 2022). In addition, the transcription level can be regulated by deleting the cis-regulatory elements of the B gene. (C) Knockout of the negative transcription factor in plant. The expression of lipid genes can be reduced in a normal plant by a variety of negative transcription factors. However, if the negative transcription factor is disrupted using CRISPR, the expression of the lipid gene can be increased. (D) CRISPR/Cas9-based technology. Epigenetic study of lipid gene seems possible if Cas9-based technique is used. For example, by methylation through dCas9-DNMT3A, lipid gene expression can be suppressed whereas demethylation via dCas9-TET can increase lipid gene expression. Alternatively, it is possible to develop a plant that slightly weakens the function of a specific protein by using base editing and has a normal phenotype than that of knockout mutants, but with a changed lipid composition.