Fig. 3.
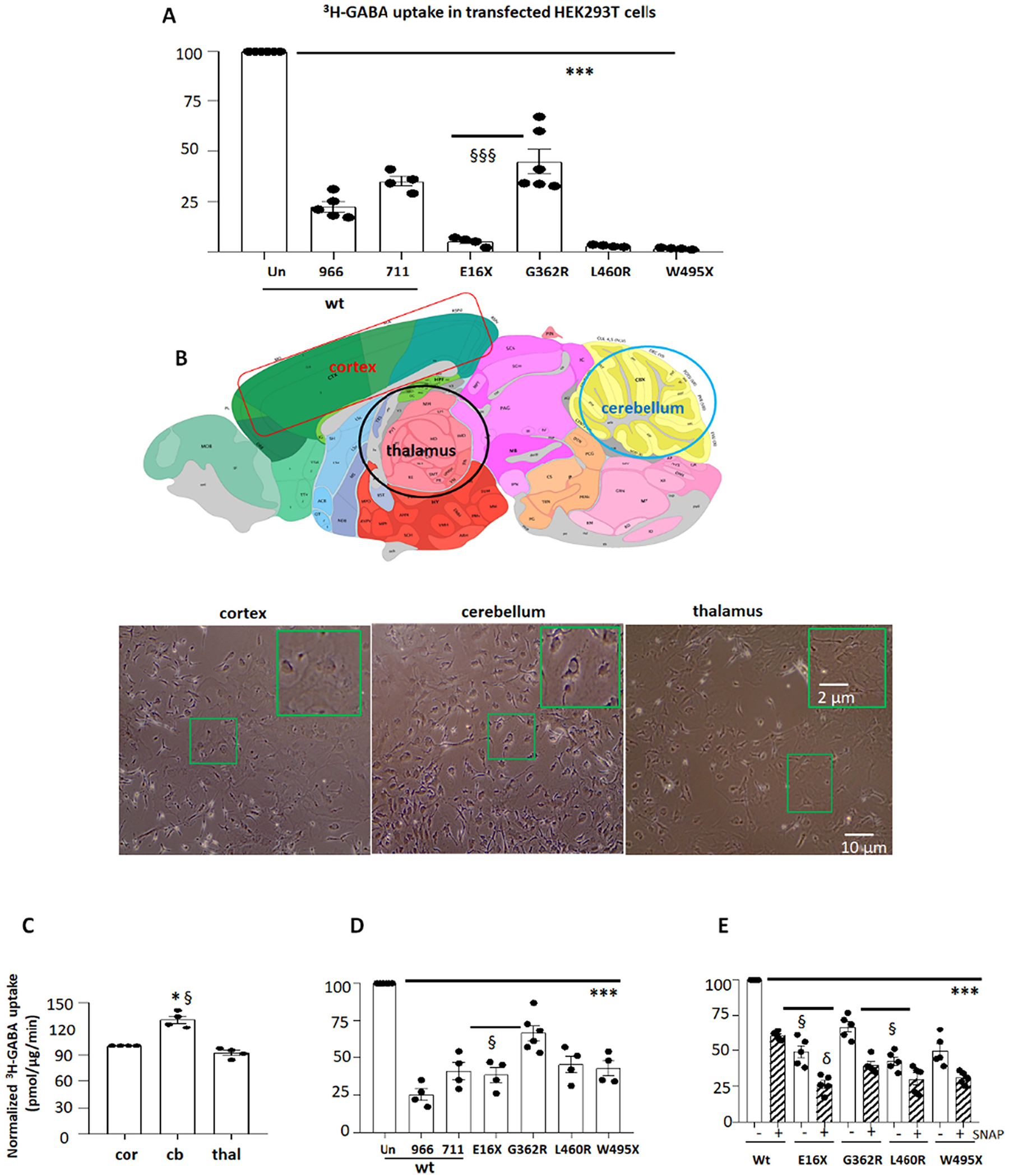
The GABA reuptake function of the variant GAT-1 transporters was reduced in HEK 293 T cells and mouse astrocytes.
(A). HEK293T cells under passage 20 were transfected with wildtype (wt) or the variant GAT-1YFP cDNAs (1 μg per a 35 mm dish) for 48 h before 3H radioactive GABA uptake assay. GAT-1-specific inhibitors Cl-966 (50 μM) and NNC-711 (30 μM) were added during flux time for each experiment. (B). Diagram adapted from the Allen Brain Atlas, demonstrating the areas that were dissected for astrocyte culture. The astrocytes from the cortex, cerebellum, and thalamus of wildtype C57BL/6 J mouse pups at postnatal day 0–3 were prepared and the live cells at passage 2 were imaged before experiment of GABA uptake assay. (C). The graph showing relative GABA uptake assay in astrocytes from cortex, cerebellum and thalamus. The GABA uptake from cerebellum (cb) and thalamus (thal) was normalized to the GABA uptake in the cortex (cor). Mouse cortical (D) or thalamic (E) astrocytes at passage 2 were transfected with wildtype (wt) or the variant GAT-1YFP cDNAs (1 μg per a 35 mm dish) for 48 h before 3H radioactive GABA uptake assay. In C, GABA uptake in astrocytes from the cerebellum and thalamus was normalized to that from the cortex (=100%) each time. In D, GABA uptake in astrocytes expressing the variant or wildtype cDNAs treated with GAT-1 inhibitors Cl-966 or NNC-711 was normalized to the astrocytes expressing the wildtype cDNAs without GAT-1 inhibition. “Un” stands for the astrocytes transfected with the wildtype GAT-1YFP without treatment of GAT-1 inhibitors. (966 stands for Cl-966 while 711 for NNC-711). In E, GABA uptake in astrocytes expressing the variant or wildtype cDNAs treated with GAT-3 inhibitor (SNAP5114 (30 μM)) was normalized to that in astrocytes expressing the wildtype cDNAs without GAT-3 inhibition. GAT-3 inhibitor was applied 1 h before and during uptake assay. In C, D and E, N = 4–5 transfections from 4 batches of cultured astrocytes, ***P < 0.001 vs astrocytes expressing the wildtype GAT-1YFP. In A, §§§P < 0.001 vs G362R. In C, *P < 0.05 vs cortex; §P < 0.05 vs thalamus. In D, §P < 0.05 vs G362R. In E, §P < 0.05 vs G362R without treatment; δ P < 0.05 vs G362R with SNAP5114 treatment. In C, D and E, N = 4–5 experiments, cells were prepared from 3 different litters of mouse pups. One-way analysis of variance (ANOVA) and Newman-Keuls test was used to determine significance compared to the wt condition and between mutations. Values were expressed as mean ± S.E.M.
