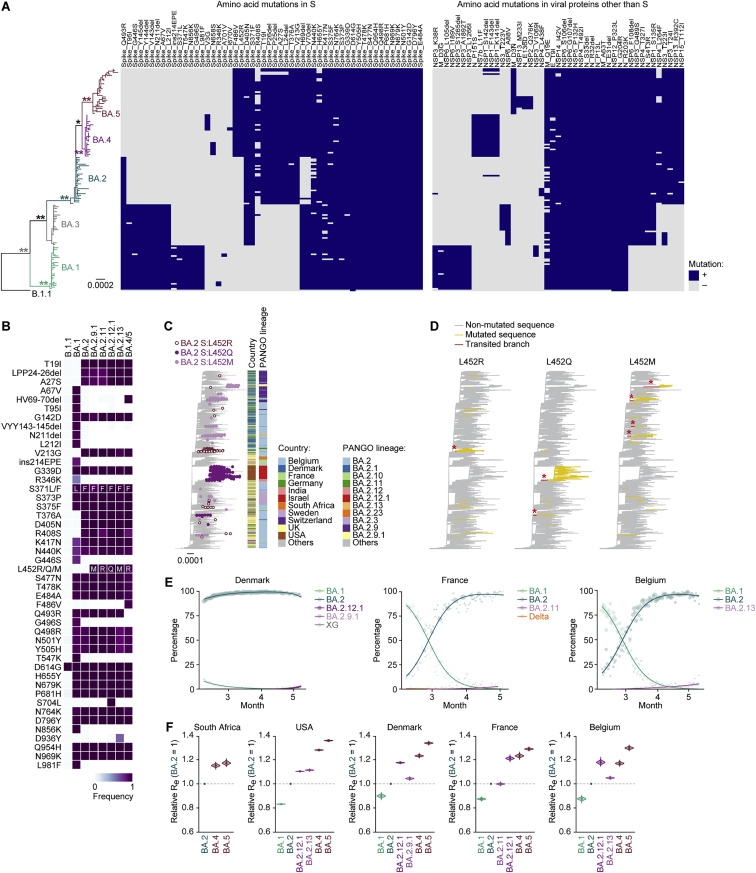
Figure S1.
Phylogenetic analysis of BA.2 subvariants, related to Figure 1
(A) The mutation profile of the Omicron lineages in South Africa, related to Figure 1A. Mutations detected in ≥5 sequences in the ML tree are summarized.
(B) Comparison of mutations in S protein among BA.2 subvariants. Mutations detected in ≥50% sequences of at least one lineage are summarized.
(C) The country and PANGO lineage of the BA.2 sequences in the ML tree, related to Figure 1B.
(D) Estimation of each common ancestry group of the S protein L452 substitution-bearing BA.2 variants. The amino acid at position 452 in the S protein in each ancestral node was estimated by a Markov model, and the branches where the L452 substitution was acquired (red branches with asterisks) were estimated.
(E) Epidemic dynamics of SARS-CoV-2 lineages. The results for up to five predominant lineages in Denmark (left), France (middle), and Belgium (right) where the BA.2-related Omicron variants bearing the S protein L452R/Q/M substitution circulating are shown. The observed daily sequence frequency (dot) and the dynamics (posterior mean, line; 95% CI, ribbon) are shown. The dot size is proportional to the number of sequences. The BA.2 sublineages without substitution at the L452 residue of the S protein are summarized as “BA.2.”
(F) Estimated relative Re of each viral lineage, assuming a fixed generation time of 2.1 days. The Re value of BA.2 is set at 1. The posterior (violin), posterior mean (dot), and 95% Bayesian confidence interval (CI) (line) are shown. Unlike Figure 1D, the SARS-CoV-2 genome surveillance data downloaded on July 7, 2022, was used.