Figure 10.
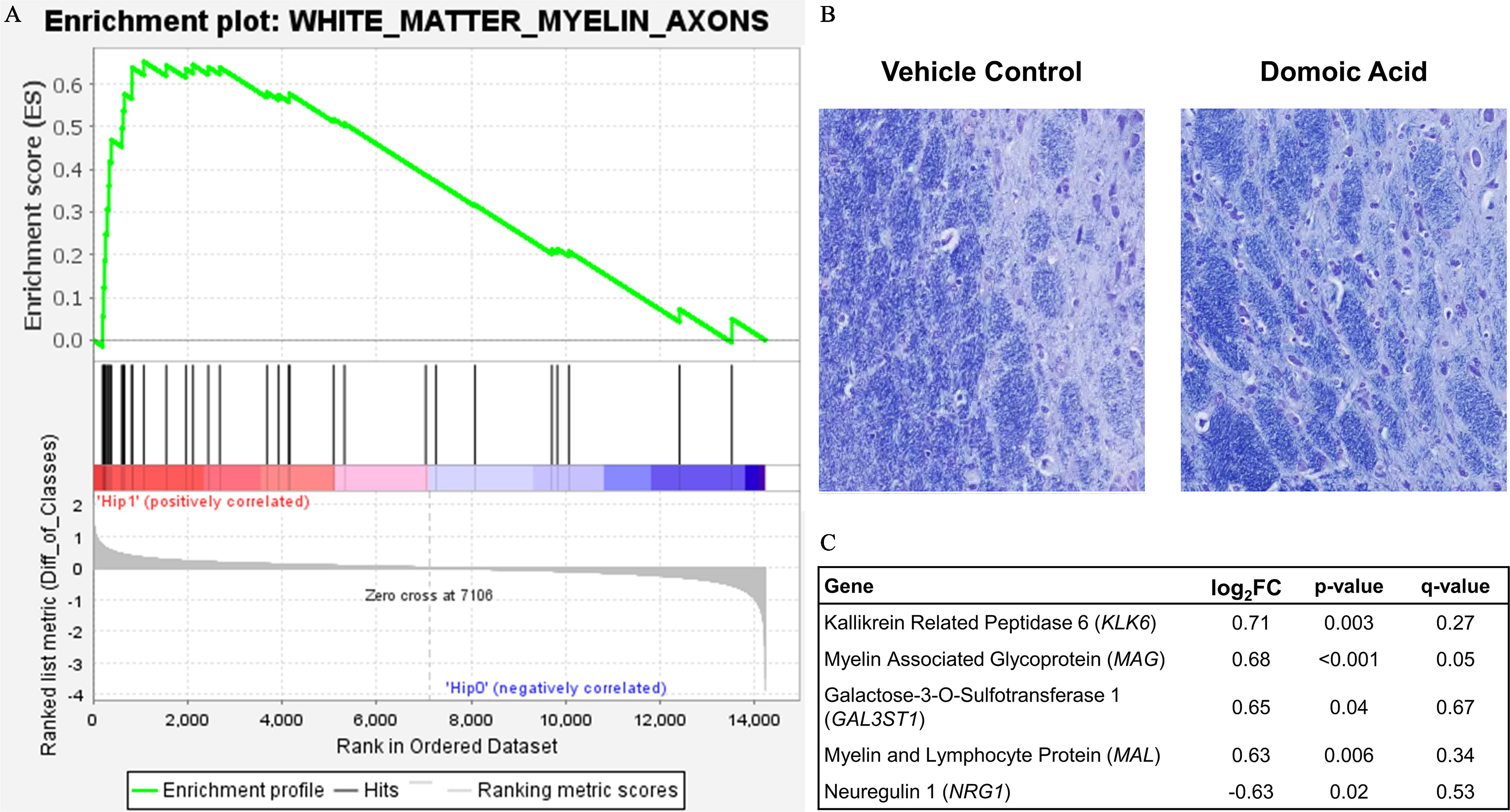
White matter response to domoic acid. (A) Gene Set Enrichment Analysis (GSEA) plot. The enrichment profile across all genes in that set from GSEA is shown in the upper panel curve, with the significant genes demarked underneath by black vertical lines and a scale representing genes up-regulated on the left (red) to those down-regulated on the right side (blue) of the panel. (B) Representation image of Luxol fast blue (LFB) staining for myelin in the hippocampus of vehicle control and domoic acid-exposed animals. (C) Significantly differentially expressed genes from the white matter GSEA list by FC. Note: Diff, difference; FC, fold change.
