Figure 9.
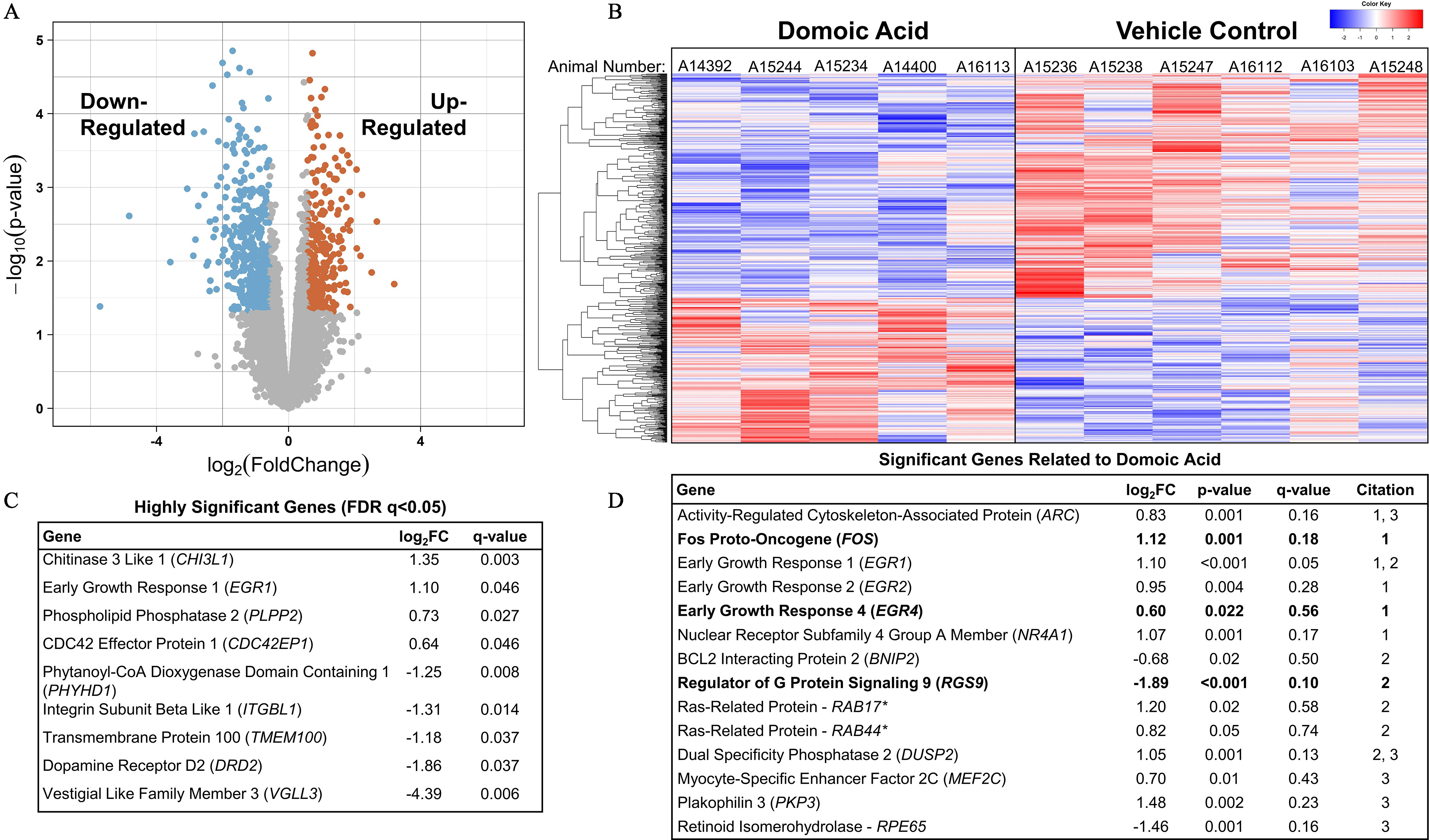
RNAseq transcriptional profiling of the hippocampus from female Macaca fascicularis following prolonged exposed to domoic acid (DA) or vehicle (5% sucrose). (A) Volcano plot shows differential expression between DA-exposed and unexposed groups. Each dot represents one gene. As indicated by the text, significantly down-regulated genes are highlighted on the left side and up-regulated gene are highlighted on the right side ( and ). Genes along the middle indicate that expression was not significantly changed. (B) Heatmap shows significantly differentially expressed genes in both the exposed animals (left) and control animals (right). Each column represents an individual animal. A14392, A15244, and A15234 were in the group, and A14400 and A16113 were in the group. Blue highlighting indicates lower expression level and is observed in the upper two-thirds of the distribution for DA-exposed animals as compared with the lower one-third of the vehicle control animals. The red indicates high expression level and is an inverse distribution to those genes showing lower expression levels. The key in the upper right corner of the image provides the density gradient for the blue (lower) and red (upper). (C) Significant genes with a false discovery rate (FDR) of . All genes in this table had a . (D) Significant genes previously reported to be differentially expressed in either a) mice,74 b) acute zebrafish,75 or c) chronic zebrafish76 after DA exposures. Bolded values indicate genes that were differentially expressed in the same direction across the previous studies. *Denotes unspecified homologous RAB gene identified in zebrafish. Note: FC, fold change; RNAseq, RNA sequencing.
