Abstract
Spheroidal cancer microtissues are highly advantageous for a wide range of biomedical applications, including high-throughput drug screening, multiplexed target validation, mechanistic investigation of tumor–extracellular matrix (ECM) interactions, among others. Current techniques for spheroidal tissue formation rely heavily on self-aggregation of single cancer cells and have substantial limitations in terms of cell-type-specific heterogeneities, uniformity, ease of production and handling, and most importantly, mimicking the complex native tumor microenvironmental conditions in simplistic models. These constraints can be overcome by using engineered tunable hydrogels that closely mimic the tumor ECM and elucidate pathologically relevant cell behavior, coupled with microfluidics-based high-throughput fabrication technologies to encapsulate cells and create cancer microtissues. In this study, we employ biosynthetic hybrid hydrogels composed of poly(ethylene glycol diacrylate) (PEGDA) covalently conjugated to natural protein (fibrinogen) (PEG-fibrinogen, PF) to create monodisperse microspheres encapsulating breast cancer cells for 3D culture and tumorigenic characterization. A previously developed droplet-based microfluidic system is used for rapid, facile, and reproducible fabrication of uniform cancer microspheres with either MCF7 or MDA-MB-231 (metastatic) breast cancer cells. Cancer cell-type-dependent variations in cell viability, metabolic activity, and 3D morphology, as well as microsphere stiffness, are quantified over time. Particularly, MCF7 cells grew as tight cellular clusters in the PF microspheres, characteristic of their epithelial morphology, while MDA-MB-231 cells displayed elongated and invasive morphology, characteristic of their mesenchymal and metastatic nature. Finally, the translational potential of the cancer microsphere platform toward high-throughput drug screening is also demonstrated. With high uniformity, scalability, and control over engineered microenvironments, the established cancer microsphere model can be potentially used for mechanistic studies, fabrication of modular cancer microtissues, and future drug-testing applications.
Keywords: tissue engineering, breast cancer, tumor modeling, biomaterials, microfluidics, drug testing
Introduction
The development of three-dimensional (3D) cancer models has paved the way for accelerated design, discovery, and validation of potential targets and drug compounds, thereby significantly reducing the time, cost, and labor involved in the drug discovery pipeline. 3D spheroidal cell aggregate models have been established as more accurate and predictive than two-dimensional (2D) models for cancer drug screening. However, current commercial techniques for spheroidal tissue formation rely heavily on the self-aggregation of single cancer cells and have substantial limitations with regard to production consistency, control, and flexibility.1−3 Although better than 2D cell models, current self-aggregation systems are unable to replicate key features of the native tumor microenvironment (TME), particularly due to a lack of control over surrounding extracellular matrix (ECM) components and heterogeneity in shape, size, and aggregate forming tendencies.2,4 Hence, there is a pressing need for novel, improved, and predictive 3D in vitro systems that enable pharmaceutical companies to obtain physiologically and pathologically relevant information on drug efficacy, safety, and response in a low-cost, facile, and high-throughput manner prior to in vivo studies and clinical trials.
3D culture of cells within biomimetic and cell-supportive engineered matrices provides physiological context to encapsulated cells through close recapitulation of native cellular microenvironments.5−9 In in vitro cancer studies, recapitulation of the complex and dynamic TME is essential for capturing various aspects of the pathological progression, including tumorigenic initiation, proliferation, invasion, and migration, among others.10−12 Tissue-engineered tumor-mimetic in vitro platforms have become physiologically relevant in recent years, providing close recapitulation of the TME.10,13 However, many of these engineered tumor platforms are not scalable for use in multiwell-plate drug-testing assays.14,15 In this regard, microfluidic fabrication technologies can be used to integrate the advantages of spheroid drug screening platforms with tissue engineering tool sets to control/manipulate the cellular microenvironment.
Developing in vitro engineered TMEs requires several important design considerations, including cancer cell type(s) and supporting cells used for encapsulation, the choice of biomaterial to be used as scaffolds/matrices,16,17 and the biofabrication technique(s) for generating synthetic cancer tissue constructs.10,13,18 The choice of the biofabrication technique is dictated by several factors including the size and shape of the final tissue construct(s), scalability and throughput, repeatability, and fabrication resolution.19,20 Encapsulation of cells within millimeter-scale hydrogel constructs presents certain challenges including (1) macromolecular mass transfer resistance over large diffusion distances; (2) uncontrolled and undesired heterogeneities in cell viability, behavior, and function; (3) low scalability, low throughput, and large volumes of cells and materials required per assay/experiment.21,22 Hence, microencapsulation technologies that enable rapid and facile creation of a large number of uniform microtissue constructs are gaining rapid attention.20 Some of these techniques include droplet microfluidics,23−28 emulsion-based1,29 and spray-based droplet generation,30−32 flow lithography,33,34 microdrop printing,35 among others. The majority of these techniques yield hydrogel microparticles, microgels, or microbeads of size ranging from submicron scale to a few hundred microns.36,37
In particular, droplet-microfluidic-based methods have been used for encapsulation of cancer cells within hydrogel microgels for the evaluation of anticancer therapies.27,38−41 Yu et al. described the automated formation of alginate microgels (of diameter ∼250 μm) encapsulating LCC6/Her-2 breast cancer cells for the assessment of doxorubicin toxicity in comparison to the standard 2D culture.27 Sabhachandani et al. reported microfluidic production of alginate-based hydrogel microspheres containing lymphoma cells, fibroblasts, and lymphocytes for simulating heterogeneous TMEs and evaluation of the immunomodulatory drug lenalidomide.38 Interestingly, continuous dynamic perfusion is applied to the cancer microgels during culture; this capacity for extended culture time is an important requirement for drug testing. Yu et al. used a microfluidic system to form breast cancer (MCF7) hydrogel microspheres with core (collagen/Matrigel) and shell (alginate) structure for the assessment of two anticancer drugs, docetaxel and tamoxifen.39 To move toward the use of cancer microgels in high-throughput screening, additional steps are needed. Upper limits on microgel size in most studies using droplet-microfluidic-based methods have been dictated by the use of photolithography, which limits the microfluidic chip outflow track dimensions and length; using these approaches, often only a small number of cancer cells per microgel have been able to be incorporated, which limits the available range in which responses can be detected by downstream assays. Additionally, most cancer microencapsulation studies, although demonstrating validation of the microgel platforms using anticancer drugs, have not yet tested compatibility with automated liquid handling, which is needed for integration with existing high-throughput screening (HTS) assays. Moreover, employing droplet-microfluidics-based methods to create engineered cancer microtissues using biomaterials which more closely recapitulate the TME may enhance the ability to obtain clinically relevant drug efficacy data from metastatic and invasive cancer cell lines.17
As an important consideration in engineering the TME, the selection of the biomaterial/biopolymer/hydrogel used for cancer cell encapsulation determines 3D cellular behavior, spatio-temporal evolution in the engineered niche, and dynamic cell–cell and cell–matrix interactions.16,17 A wide range of natural, synthetic, and hybrid hydrogels have been used for 3D cancer studies.7,17,42,43 Alginate, although advantageous for use in droplet-microfluidic-based microgel production, does not inherently support recapitulation of characteristic tumor cell morphologies in 3D culture. Hybrid hydrogels comprising a natural component that supports cell adhesion and matrix remodeling (e.g., protein or protein-derived peptide sequence) and a synthetic component that enables rapid crosslinking and tuning of matrix properties (e.g., crosslinked polymeric macromolecular chains) provide an optimal approach for exploiting the advantages of both natural and synthetic materials.5,17 In this study, we employ a hybrid hydrogel, poly(ethylene glycol) (PEG) diacrylate (PEGDA) covalently conjugated with fibrinogen to form PEG-fibrinogen (PF), for establishing an in vitro engineered cancer microtissue platform. PF hydrogels have been previously used in 3D in vitro cancer studies for the evaluation of cancer cell–matrix interactions in response to tuned matrix compositions, tumorigenic progression, and drug-efficacy testing.1,21,44−46
Using PF as the base precursor, we aimed to generate hydrogel microspheres in the sub-millimeter range for potential application in HTS assays and ease of robotic handling. A facile droplet microfluidics approach with custom-modified design (photolithography-free) and operation parameters was adopted for efficient and consistent generation of hydrogel microspheres encapsulating cancer cells (cancer microspheres) and their subsequent 3D culture and characterization.44,45 PF hydrogel-based uniform monodisperse cancer microspheres of diameter 800–1000 μm were generated in a rapid, high-throughput manner. Two breast cancer cell lines (MCF7, weakly metastatic; and MDA-MB-231, highly invasive and metastatic) were encapsulated, maintained in 3D culture, and characterized in terms of viability, metabolic activity, proliferation, 3D morphology, and stiffness over 14 days in culture. Cancer cells demonstrated cell-type specific differences in 3D morphology, proliferation, and metabolic activity, thereby demonstrating the ability of the PF microsphere platform for conducting mechanistic cancer studies. Finally, proof-of-concept drug testing was conducted on these cancer microspheres by shipping them to an HTS laboratory to demonstrate their translatability toward HTS assays. The cancer microsphere platform developed in this study can be potentially utilized both for mechanistic studies of the TME, tumor progression, and tumor–matrix interactions, as well as for validation studies in the drug discovery process.
Materials and Methods
Cell Culture and Maintenance
MCF7 (ATCC HTB-22) and MDA-MB-231 (ATCC HTB-26) human breast cancer cells were obtained from ATCC (Manassas, VA). Cancer cells were maintained in Dulbecco’s Modified Eagle’s Medium (Gibco) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals), 1% (v/v) nonessential amino acids (NEAAs) (Lonza), 1% (v/v) penicillin/streptomycin, 1% (v/v) glutamax (Gibco), and 1% (v/v) sodium pyruvate.
Hydrogel Synthesis and Characterization
Poly(ethylene glycol) diacrylate (PEGDA) was prepared as described previously.47 Bovine fibrinogen was covalently coupled to PEGDA according to previously established protocols.1,21 In brief, fibrinogen was dissolved in a phosphate-buffered saline (PBS) buffer with 8 M urea at a concentration of 7 mg/mL. Tris (2-carboxyethyl)phosphine hydrochloride (TCEP-HCl) was added at a molar ratio of 1.5:1 TCEP to fibrinogen cysteines to the above solution, and the final pH was adjusted to 8.0. PEGDA was dissolved in the urea–PBS buffer at 280 mg/mL, and the solution was slowly added to the fibrinogen solution. The reaction was allowed to proceed for 3 h at 25 °C in the dark. The solution was then diluted with an equal volume of urea–PBS buffer and precipitated in acetone at a volumetric ratio of 4:1 of acetone to product solution. The precipitate was separated by centrifugation, weighed, and redissolved in urea–PBS buffer at 2.2 mL of buffer/g of the precipitate. The product was dialyzed against sterile PBS over 24 h (with three changes of PBS) at 4 °C in the dark. The final product was aliquoted into sterile centrifuge tubes and stored at −80 °C.
Synthesized PF was characterized for fibrinogen concentration using a standard Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Scientific) with bovine serum albumin (BSA) as standards. PEGDA concentration was determined by aliquoting a known volume of PF solution in glass vials, lyophilizing, and measuring the net weight of the dry solid obtained.
Cell Encapsulation in PF Microspheres
Cell encapsulation in PF hydrogel microspheres was achieved through a microfluidic poly(dimethyl siloxane) (PDMS) device with a modified T-junction design.44,45 A PDMS mold was created using a Sylgard 184 silicone elastomer kit (Dow Corning) by pouring the mixture of the base and cure components (ratio of 10:1) into a 3D printed bracket containing the preassembled microfluidic channel mold. The mixture was subsequently degassed and heat-cured at 70 °C for 2 h. After curing, the microfluidic channel mold was disassembled. The microfluidic PDMS mold was sonicated in 70% ethanol for cleaning and sterilization before and after each use.
Before cell encapsulation, the polymer precursor solution was prepared by mixing PF with 0.1% (w/v) of Pluronic F68 (Sigma), 0.1 mM eosin Y photoinitiator (Fisher Scientific), 1.5% (v/v) of triethanolamine (TEOA) (Acros Organics), and 0.39% (v/v) of N-vinyl pyrrolidinone (NVP) (Sigma). Cancer cells cultured in tissue-culture flasks were trypsinized and resuspended in the hydrogel precursor at 20 × 106 cells/mL.
The PDMS microfluidic T-junction had two inlets and one outlet; the cell-laden polymer precursor was injected from the top inlet at 1 mL/h, mineral oil was injected from the bottom inlet at 10 mL/h via syringe pumps, and crosslinked microspheres were collected at the outlet. When the two phases interfaced at the T-junction, the aqueous polymer phase was pinched into droplets by the oil phase and carried along the channel. Cell-encapsulated microspheres were crosslinked by a broad-spectrum visible-light source (intensity: 2.8 W/cm2) (Prior Lumen 200). A mirror was placed behind the microfluidic channel within a distance of approximately 0.5 cm to aid the crosslinking by reflecting the transmitted light near the zone of crosslinking. The microspheres were washed down from the outlet with prewarmed media injected via a third syringe pump set at 22 mL/h. The microspheres were centrifuged at 200 g for 3 min, and the supernatant oil layer was removed. The oil-free microspheres were resuspended in media and transferred to well plates for 3D culture over 14 days.
Quantification of Microsphere Size and Distribution
Phase-contrast images of cell-laden microspheres were obtained at defined time points using a Nikon Ti inverted microscope equipped with an Andor Luca S camera. The microsphere diameter was analyzed in ImageJ software (NIH, Version 1.52n) by manually drawing a region of interest around the microsphere edges. A minimum of 50 microspheres were analyzed per batch.
Assessment of Cell Viability
Cell viability within PF hydrogel microspheres was assessed at defined time points by the Live/Dead viability kit (Invitrogen) as per the manufacturer’s protocol. Live cells were stained with calcein AM, and dead cells were stained with ethidium homodimer 2. Fluorescence images of cell-laden microspheres were obtained via a confocal microscope (Nikon AI confocal scanning laser microscope), and images were analyzed in ImageJ software to quantify cell viability. Each image stack was analyzed slice by slice, and the number of live cells (labeled green) and dead cells (labeled red) were manually counted. Cell viability was quantified as (number of live cells)/(number of live cells + number of dead cells). A minimum of three microspheres were analyzed for each time point and each cell type.
Assessment of Microsphere Elasticity
Elastic moduli of acellular and cell-laden microspheres were assessed via a parallel-plate compression testing system. Fabricated microspheres were loaded onto the CellScale Microsquisher platform, maintained at 37 °C in PBS, preconditioned for compression testing, and made to undergo cycles of compression and relaxation at a rate of 5 μm/s for a minimum of 15% strain. The force–displacement data obtained from the compression test (via SquisherJoy software) were converted to stress–strain curves, and the lower portion of the curve (5–15% strain) was used to estimate the elastic moduli of the microspheres. A minimum of three microspheres were measured for each condition.
Assessment of Metabolic Activity
The metabolic activity of cell-laden microspheres was assessed using the 2,3-bis-(2-methoxy-4-nitro-5-sulphenyl)-(2H)-tetrazolium-5-carboxanilide (XTT) assay (Biotium) according to the manufacturer’s protocol. Cell-laden microspheres were transferred into a 96-well-plate with one microsphere per well in 100 μL of media. 25 μL of the XTT working solution was added to each well. After incubation for 18 h at 37 °C, absorbance (470 nm) and background (660 nm) intensities were measured via a microplate reader (Bio-Tek). The background intensity was subtracted from the absorbance intensity to obtain the relative absorbance intensity for each microsphere. A minimum of five microspheres were analyzed per condition at each time point. The relative metabolic rate obtained was normalized to day 0 values for each cell type.
Ultrastructural Visualization
The ultrastructural features of cell-laden microspheres were visualized through scanning electron microscopy (SEM). Cell-laden microspheres were washed with PBS, fixed with 4% glutaraldehyde for 1 h, and postfixed with 2% osmium tetroxide (electron microscopy sciences, EMS) for 1 h, all at 25 °C. The fixed microspheres were dehydrated gradually in increasing concentrations of ethanol and chemically dried using hexamethyl disilazane (HMDS) (EMS) for 3 h. Dried samples were mounted on carbon-taped aluminum stubs, sputter-coated with gold (Pelco SC-6 sputter coater), and imaged using a scanning electron microscope (JEOL JSM-7000F).
Immunofluorescence Staining and Imaging
The 3D morphology and proliferation of cell-laden microspheres were visualized by immunostaining and confocal fluorescence microscopy. Cell-laden microspheres (post-14 days in 3D culture) were washed with PBS, fixed with 4% paraformaldehyde for 1 h at 25 °C, permeabilized with 0.5% Triton-X for 30 min, followed by incubation with a blocking buffer (2% BSA and 5% FBS in PBS) overnight. On the following day, the constructs were incubated with the primary antibody for Ki67 (human antirabbit, Abcam, 1:100 dilution) for 3 h, followed by incubation with the secondary antibody for Ki67 (Alexa Fluor 488 goat antirabbit, Invitrogen, 1:200 dilution) and Alexa Fluor 568 Phalloidin (Invitrogen, 1:200 dilution) for 3 h. Finally, the microspheres were stained with Hoechst 33342 (1:200 dilution) for 1 h and washed with PBS. Stained microspheres were mounted on coverslips and imaged using confocal microscopy to obtain z-stacks. Due to limitations of the objective working distance, z-stacks of microspheres were limited to 300 μm in height.
Assessment of Drug Efficacy
Cell-laden microspheres were produced in Dr. Lipke’s laboratory at Auburn University, Alabama, and shipped to Dr. Minond’s laboratory at Nova Southeastern University, Fort Lauderdale, Florida, overnight for drug efficacy assessment. The microspheres were kept in sealed centrifuge tubes filled completely with the medium and shipped at ambient temperature. The packaging was prepared following regulations for UN3373 biological substances category B. Maintenance of cell viability during shipping was confirmed after the package was delivered.
To test the effect of known chemotherapeutic drugs on MDA-MB-231 cell-laden microspheres, 100 μM doxorubicin was incubated with the microspheres in 384-well plates. Viability was tested after 48 h using the Live/Dead viability kit (Invitrogen) and imaged on a CellInsight CX7 high-content imaging platform (Thermo Fisher Scientific). Additionally, 384-well plates containing MDA-MB-231 microspheres were treated with 1 μM staurosporine and 10 μM doxorubicin using an automated liquid handler (Biomek NXP Beckman Coulter). Viability of the cells was measured after 72 h using a CellTiter-Glo 3D luminescent cell viability assay (Promega) using a Bio-Tek Synergy microplate reader (Bio-Tek Inc., VT, US).
Statistical Analysis
All statistical analysis was performed using Minitab 17 Statistical software (Minitab Inc.). After checking for normality of distribution, one-way ANOVA with Tukey’s family error rate of 5% was used to evaluate the statistical significance between multiple groups, assuming equal variance and equal sample size of compared groups. Unless otherwise indicated, p < 0.05 was considered statistically significant.
Results and Discussion
Fabrication and Size Characterization of Cancer Microspheres
In order to consistently fabricate uniform PF hydrogel microspheres, a microfluidic system employing an aqueous-oil dual-phase system was established. Poly(dimethyl siloxane) (PDMS) was used to construct a modified T-junction mold with an outlet channel diameter of 900 μm and a length of 10 cm. The two inlet ends of the T-junction were connected to syringes containing the cell-laden PF hydrogel precursor (syringe pump 1 connected to top inlet) and mineral oil (syringe pump 2 connected to bottom inlet), respectively. The top aqueous inlet comprised a restriction segment for stabilization of the aqueous/oil interface. The bottom inlet of the T-junction comprised a tapering end to eliminate the dead volume of the hydrogel precursor prior to entering the outlet. The outlet also featured a tapering end at the T-junction to increase the flow speed during droplet formation. The vertical orientation of the T-junction, as opposed to a horizontal orientation found in other studies, improved the separation efficiency of the denser aqueous phase by the lighter oil phase. These modified T-junction design features, adopted from flow-focusing strategies, were critical for obtaining rapid aqueous-phase droplet formation at optimized flow rates.
The composition of the hydrogel precursor and oil phase was determined based on previous studies using dual-phase emulsion techniques to create hydrogel microspheres.1 After chemical conjugation of PEGDA and fibrinogen, the yield and PEGylation efficiency were estimated and found to be similar to those reported previously.21 The degree of acrylation of PEGDA, measured through proton NMR, was reported as 96%. The PF obtained after dialysis contained 16.6 mg/mL fibrinogen (determined by the BCA assay) and 21.5 mg/mL PEGDA (determined by measuring the dry weight after lyophilization). The mass of the total polymer was 3.8% w/v, and the molar ratio of PEGDA: fibrinogen was 44:1. The PF hydrogel precursor in PBS was mixed with eosin Y as the visible-light-based photoinitiator, TEOA as the coinitiator, NVP as the comonomer, and Pluronic F68 as the aqueous-phase surfactant to stabilize the formed droplets. Cancer cells were resuspended in the precursor at 20 × 106 cells/mL. The initial cell encapsulation density within the hydrogel precursor can be varied from 5 × 106 to 60 × 106 cells/mL without affecting the photo-crosslinking efficiency of the polymer precursor or uniformity of hydrogel microsphere size distribution. A broad-spectrum light source was positioned midway over the channel with a mirror being positioned behind the channel for reflecting and focusing light within a distance of approximately 0.5 cm for rapid crosslinking (crosslinking time: ∼1–2 s) (Figure 1A). Shielding of the T-junction from incident light was necessary to prevent undesired crosslinking at the T-junction and clogging of the microfluidic channel. Optimization of the aqueous-phase flow rate to 1 mL/h and oil-phase flow rate to 10 mL/h resulted in uniformly crosslinked microspheres in the size range of 750–950 μm in diameter. The end of the microfluidic channel was connected to a third syringe pump containing media to collect and wash off crosslinked microspheres and to prevent them from accumulating at the channel end. The cell-laden PF microspheres in media were centrifuged, and the supernatant oil layer was aspirated, after which the microspheres were resuspended in media and maintained in 3D culture over 14 days.
Figure 1.
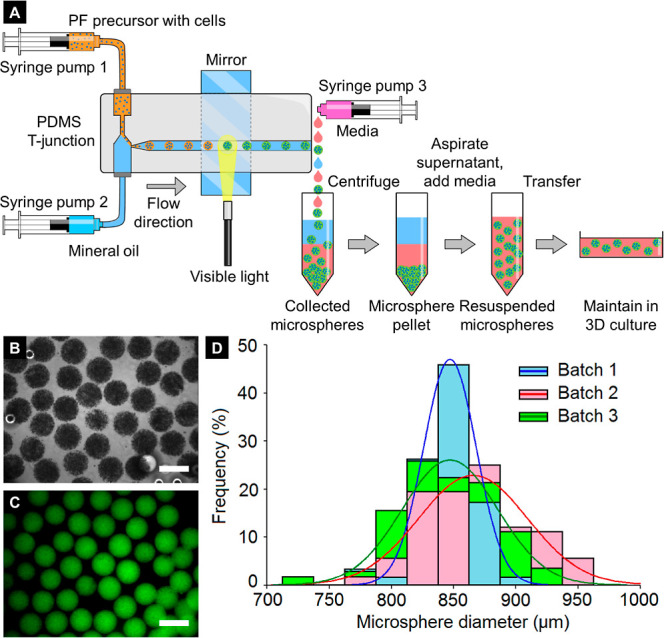
Microfluidic fabrication and size characterization of cell-laden PF microspheres. (A) Schematic of the microfluidic setup for the generation of cell-laden PF microspheres and 3D culture. (B) Phase-contrast image of MCF7 cell-encapsulated microspheres. (C) MCF7 microspheres imaged under green fluorescence (due to the presence of the eosin Y photoinitiator in the hydrogel precursor). Scale bar = 1000 μm. (D) Distribution of microsphere diameter (with normal fits) from three independent batches demonstrates the degree of interbatch and intrabatch uniformity and monodispersity, n = at least 50 microspheres per batch.
Phase-contrast and fluorescence visualization of crosslinked microspheres revealed an overall uniform appearance and size distribution of microspheres. Encapsulated cells were also observed to be uniformly distributed within individual microspheres (Figure 1B,C). Further quantification of microsphere size immediately postfabrication revealed a high degree of interbatch and intrabatch uniformity and a low coefficient of variation (CV) with the majority of microspheres within 800–900 μm in diameter [batch 1: diameter = 850 ± 20 μm (mean ± standard deviation), CV = 2.4%; batch 2: diameter = 870 ± 40 μm, CV = 4.6%; batch 3: diameter = 850 ± 40 μm, CV = 4.7%] (Figure 1D).
Microfluidic fabrication of monodisperse micron-scale hydrogel constructs has previously been executed via a number of techniques including continuous-flow lithography (CFL),34 stop-flow lithography (SFL),33 parallelized step emulsification (PSE),48 flow focusing and droplet emulsification (FF-DE),23−25,49−51 among others. Each of these techniques has its own advantages and limitations, which make them conducive toward specific applications. Some of these applications include modular synthesis and assembly of microtissue constructs,48,51 cell encapsulation for therapeutic cell delivery,49,50,52 vascularization of macroporous hydrogel assemblies,53 and drug delivery.54−56 In this study, we combined the working principles of CFL (continuous flow across a projected optical path) and FF-DE (dual-phase aqueous-oil flow and hydrogel precursor droplet pinching) to create a microfluidic fabrication system that enables high-throughput generation of nanoliter volume hydrogel microspheres with large size and high cell density.
Here, we employ larger spheroidal tissues with a much high cell density than are typically produced using microfluidic systems.45,57 Using our system, the total cell number per microsphere is much greater than can be encapsulated in smaller microspheres, providing a larger dynamic range for assessing drug response, greater well-to-well reproducibility as a result of higher tolerance to cell number variances between microspheres, and arguably a more biologically relevant TME through the recapitulation of in vivo and patient tumor diffusion limitations.
Overall, the established microfluidic system was able to facilitate the high-throughput generation of uniform cell-laden PF hydrogel microspheres for subsequent 3D breast cancer cell culture and investigation of tumorigenic behavior. Additionally, this approach eliminates the need for costly microfabrication and photolithographic and cleanroom expenses and the entire setup can be easily assembled or disassembled within a standard cell culture hood space.
Assessment of Cell Viability within Cancer Microspheres
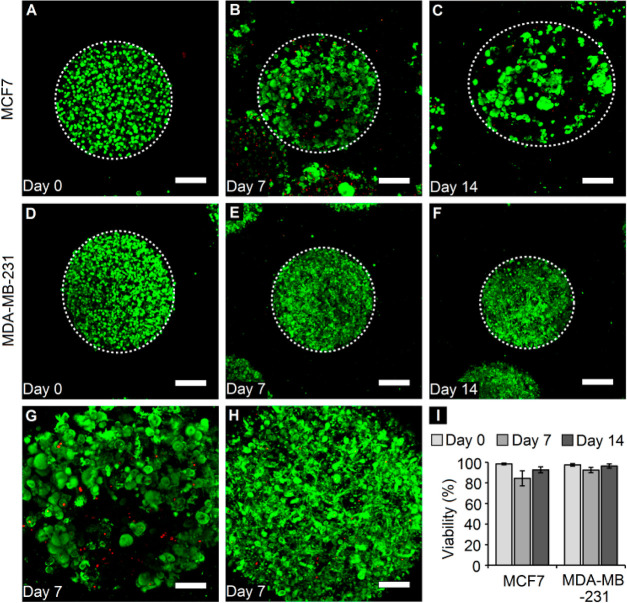
Post-microsphere fabrication and cell encapsulation, the ability to maintain breast cancer cells in 3D culture within the PF hydrogel microspheres over 2 weeks was demonstrated. The cell viability for both MCF7 and MDA-MB-231 breast cancer cells was visualized and assessed via fluorescence staining and confocal imaging on day 0 (few hours postencapsulation), day 7, and day 14 (Figure 2A–F). The cell viability for both cell types on day 0 was found to be greater than 95% (Figure 2A,D,I), indicating that the microfluidic fabrication technique (including the use of the UV light source) did not have any significant impact on cell viability. In general, high viability was maintained for both cell types through 14 days in culture (>90%), except for MCF7 cells, which displayed a slight drop in viability to 84% on day 7 (Figure 2I). Closer inspection of encapsulated cells revealed that MCF7 cells grew as local dense colonies or clusters within the microspheres (Figure 2G), while MDA-MB-231 cells appeared to spread out and occupy the void space within the microspheres without any tendency of cluster formation (Figure 2H).
Figure 2.
Cell viability within cancer microspheres. (A–C) Confocal z-projections of MCF7 cells and (D–F) those of MDA-MB-231 cells over 2 weeks in culture. White dotted circles denote the edge of microspheres. (Green: calcein AM, live cells; red: ethidium homodimer, dead cells) (scale bar = 200 μm). (G) High-magnification z-projections of MCF7 cells and (H) MDA-MB-231 reveal overall high viability and differences in the cell-type-dependent morphology (scale bar = 100 μm). (I) Quantification of cell viability within PF microspheres reveals high viability for both cell types through 14 days in culture. n = minimum 3 microspheres per time point per cell type.
MCF7 cells, being epithelial in nature, grow as tight clusters with well-formed cell–cell junctions in 2D culture. MDA-MB-231 cells, being mesenchymal in nature, grow as solitary cells with elongated morphologies and long invasive protrusions in 2D culture, indicative of their migratory potential. Similarly, in 3D culture within PF microspheres, MCF7 cells adopted a clustered morphology with tight packing within individual clusters and a gradual increase in cluster size over time. MDA-MB-231 cells, on the other hand, grew in the PF microspheres as elongated, protruding single cells indicative of their invasive morphology, even escaping the microspheres over time. These cell-type-specific differences demonstrate the suitability of the PF hydrogel microsphere platform in elucidating 3D behavior relevant to the cancer type, thereby providing biological relevance to the in vitro tumor model. Overall, the ability to encapsulate breast cancer cells within PF hydrogel microspheres and maintain them with high viability in 3D culture over time was demonstrated.
Characterization of Microsphere Diameter, Elasticity, and Cellular Metabolic Activity
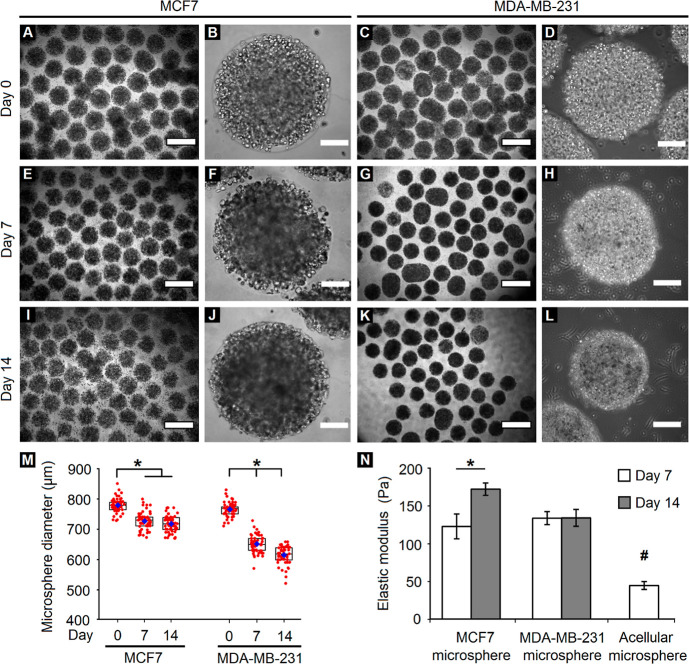
Having established the ability to maintain cancer cells within PF hydrogel microspheres, further characterization of the changes in microsphere size and stiffness over time was conducted. When visualized through phase-contrast microscopy, MCF7 cells appeared to form dense local colonies arising from single encapsulated cells; these colonies grew darker over time, indicating progressive cell growth (Figure 3A,B,E,F,I,J). In contrast, MDA-MB-231 cells appeared to form elongated morphologies within the microspheres over time. Interestingly, a sharp decrease in diameter was observed for MDA-MB-231 microspheres through 14 days in culture, possibly due to contractile stresses exerted by elongated cells on the PF hydrogel matrix. Quantification of the microsphere diameter confirmed these observations (Figure 3M). In case of MCF7 cells, the microsphere diameter significantly reduced from day 0 (average: 780 ± 20 μm) to day 7 (average: 730 ± 30 μm) and remained fairly steady thereafter. In case of MDA-MB-231 cells, the microsphere diameter progressively decreased from day 0 (average: 770 ± 20 μm) to day 14 (average: 610 ± 30 μm) of culture.
Figure 3.
Variation in microsphere size and stiffness over time. (A–L) Phase-contrast images of breast cancer cells encapsulated within PF microspheres demonstrate variations in size distribution and cellular appearance of MCF7 and MDA-MB-231 cells. (A,C,E,G,I,K: scale bar = 1000 μm; B,D,F,H,J,L: scale bar = 200 μm). (M) Quantification of the microsphere diameter reveals a significant reduction in size for both cell types over 14 days in culture (*p < 0.05, n = minimum of 50 microspheres). Red circles denote individual microspheres, blue diamonds denote the mean, while rectangular boxes represent lower quartiles, medians, and upper quartiles of the respective groups. (N) Quantification of the elastic modulus reveals a significant increase in MCF7 microsphere stiffness but no variation in MDA-MB-231 microsphere stiffness over time. The acellular microsphere stiffness is significantly low compared to that of the cell-laden microspheres for both cell types. (*,#p < 0.05, n = 3 microspheres per group).
In order to assess the effect of cell encapsulation and varying cellular morphology on the stiffness of PF hydrogel microspheres, the elastic moduli of cellular and acellular microspheres were quantified via parallel-plate compression testing. The elastic moduli of MCF7 microspheres increased significantly from day 7 (123 ± 16 Pa) to day 14 (172 ± 8 Pa), while that of MDA-MB-231 microspheres remained constant over time (∼134 ± 10 Pa). Interestingly, the elastic moduli of acellular microspheres (45 ± 5 Pa) were significantly less than those of cell-encapsulated microspheres for any given time point, indicating the influential and dominant role played by the bulk stiffness of PF hydrogel microspheres (Figure 3N).
PF hydrogels, owing to their inherent low elasticity, are susceptible to cell-generated forces and can be remodeled mechanically under biophysical forces generated by encapsulated cells, as well as by cell-responsive matrix degradation. Due to their epithelial nature, MCF7 cells grew as tight clusters within the hydrogel, expanding outwardly into the matrix with cell division. For cluster growth to occur, reorganizing MCF7 cells also likely exerted contractile forces on the matrix. Balancing these two effects, microspheres containing MCF7 cells had a relatively small, but noticeable decrease in size over 14 days in culture. In contrast, MDA-MD-231 cells, being mesenchymal in nature, spread out as single cells and have the potential to generate high traction forces to facilitate cell migration. The cumulative effect of the MDA-MB-231 cells adhering to and pulling against the surrounding hydrogel matrix to form their contractile protrusions likely contributed to the observed inward bulk shrinkage and compaction of the microspheres. Parallelly, we also observed that MDA-MB-231 cells degraded the hydrogel matrix faster than MCF7 cells, leaving the microspheres and growing in the surrounding well plate. Thus, over time, MDA-MB-231 cells become more invasive and protrusive in their morphology. The combined effect of individual cell-generated forces and matrix degradation was likely responsible for the gradual but significant shrinkage of the microspheres from ∼770 to ∼610 μm over 14 days in culture.
Further assessment of the relative changes in metabolic activity of encapsulated cells within PF hydrogel microspheres through 14 days in culture was conducted using the XTT assay (Figure 4). MCF7 cells displayed increasing relative metabolic activity through 14 days with significantly high activity on day 14, possibly due to cell proliferation and increasing local colony formation. However, the metabolic activity of MDA-MB-231 cells remained fairly constant over time, indicating potential metabolic quiescence or relatively low-proliferative behavior of this cell type within PF hydrogel microspheres. Considering that the relative cell density of MCF7 cells in the PF microspheres increased due to cluster formation and cluster expansion, a significant increase in metabolic activity over time is expected. Interestingly, MDA-MB-231 cells remained as single elongated cells within the microspheres, displaying a high degree of migratory behavior and even escaping the microspheres over time. Hence, the relative cell density of MDA-MB-231 cells could have remained fairly constant, which explains the nonsignificant changes in the metabolic activity of MDA-MB-231 microspheres.
Figure 4.
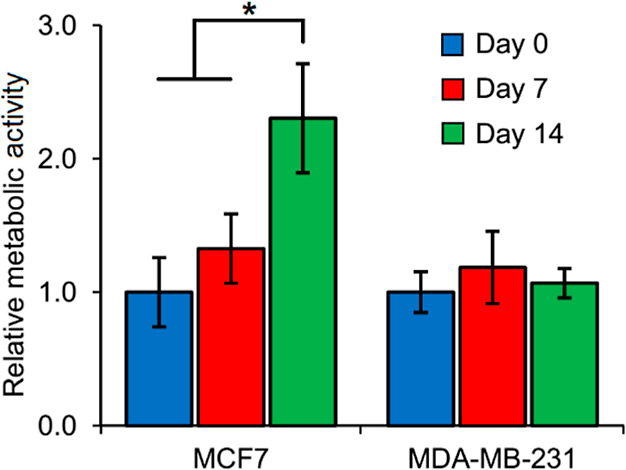
Metabolic activity of cells within cancer microspheres. MCF7 cells display increasing metabolic activity through 14 days with significantly higher activity on day 14. MDA-MB-231 cells display fairly steady metabolic activity through 14 days in culture. All values are normalized to day 0 values for each cell type. (*p < 0.05, n = 5 microspheres per group).
Overall, the encapsulation of cancer cells within PF hydrogel microspheres resulted in gradual changes in microsphere size, stiffness, and metabolic activity over time, which was modulated by the cell-type dependent morphology, 3D cell behavior, and potential cell-PF hydrogel matrix interactions.
Ultrastructure and 3D Cellular Morphology in Cancer Microspheres
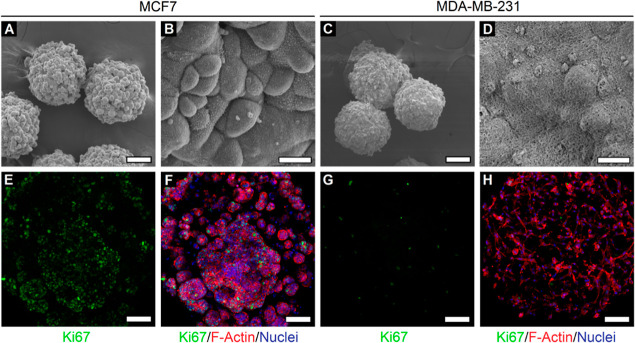
In order to visualize the ultrastructural surface features of cells encapsulated within cancer microspheres, SEM imaging was conducted. The SEM images of MCF7 microspheres revealed dense local colony formation with spherical-shaped colonies being distributed throughout the microsphere surface (Figure 5A). Closer inspection revealed the dense packing of MCF7 cells with tight cellular interactions within individual colonies and microvillus-like features present on the cell surface (Figure 5B). MDA-MB-231 microspheres displayed different ultrastructural features compared to MCF7 microspheres (Figure 5C). MDA-MB-231 cells appeared as individual cells and interspersed within the mesh-like PF hydrogel matrix (Figure 5D).
Figure 5.
Ultrastructure and 3D morphology of cells within cancer microspheres. (A) SEM image of MCF7 microspheres reveals dense local colony formation in individual microspheres. (B) At higher magnification, MCF7 cells appear to be tightly packed within individual colonies with visible cell-surface microvillus-like features. (C) SEM image of MDA-MB-231 microspheres and (D) higher magnification image revealing individual cells (bulge-like appearance) interacting with the surrounding PF hydrogel matrix. (A,C: scale bar = 100 μm; B,D: scale bar = 10 μm). Microspheres appear smaller in size due to the shrinkage effects of SEM sample preparation. (E–H) Confocal z-projection of MCF7 and MDA-MB-231 cells reveals prominent cell-type-dependent differences in proliferation and 3D morphology (green: Ki67, red: F-actin, blue: nuclei; scale bar = 200 μm). All images were obtained on day 14 of culture.
The 3D morphology and cellular arrangement of cancer cells within PF hydrogel microspheres were further visualized by fluorescence staining and confocal imaging. MCF7 microspheres revealed a higher degree of Ki67 positive staining compared to MDA-MB-231 microspheres (Figure 5E,G), indicating higher cell proliferation in MCF7 cells. This observation also correlates with the previous assessment of cellular metabolic activity in Figure 4. MCF7 cells presented a rounded morphology (as observed through F-actin arrangement) and appeared to grow as distinct local colonies with tight cell packing and distributed uniformly within the microspheres (Figure 5F). However, MDA-MB-231 cells presented elongated morphologies with a high degree of cellular projections and filopodial extensions (Figure 5H), reminiscent of their migratory and invasive behavior. These observations are in line with those reported previously in gelatin methacrylate hydrogels,58 where MCF7 cells grew as tight rounded clusters, while MDA-MB-231 cells displayed invasive protrusions and migratory behavior. In general, breast cancer cells encapsulated within PF hydrogel microspheres displayed cell-type-dependent differences in ultrastructural and morphological characteristics. These differences reveal important insights into 3D cell behavior under the synergistic influence of cell–cell and cell–matrix interactions within PF microspheres.
Demonstration of Automated Drug Testing on Cancer Microspheres
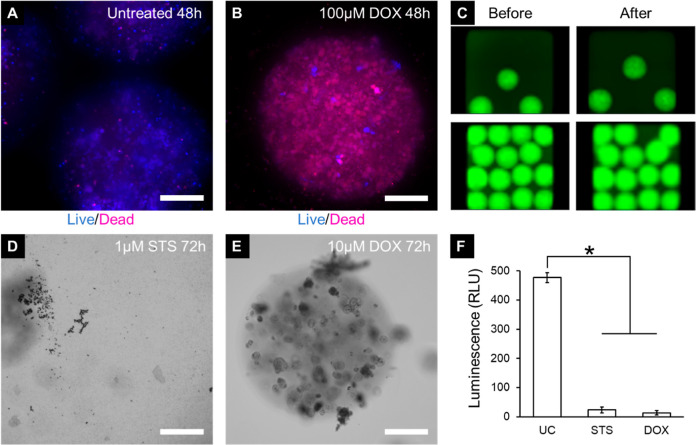
To demonstrate the feasibility of using the microspheres for high-content drug testing, MDA-MB-231 microspheres were shipped overnight to test the capacity for distribution, and maintenance of cell viability during shipping was confirmed. MDA-MB-231 microspheres were then treated with 100 μM doxorubicin, and drug-induced changes in viability were subsequently tested. Untreated microspheres exhibited a robust signal from live cells, suggesting that the cancer cells remain viable in 384-well plates for the assay duration (Figure 6A). Cells treated with 100 μM doxorubicin showed a significant loss of viability (Figure 6B). Therefore, doxorubicin administered at high concentrations is expected to provide a strong positive control for high-content assay development.
Figure 6.
Drug testing on MDA-MB-231 microspheres. (A) Untreated microspheres for control are mostly live, whereas (B) cells treated with 100 μM doxorubicin for 48 h are mostly dead (blue: live; red: dead). Fluorescent images taken using the CellInsight CX7 high-content imaging platform. (C) Images of wells containing MDA-MB-231 microspheres before (left column) and after (right column) the pinning by a 384-pin tool mounted on Biomek NXP automated liquid handler, which is used for delivering drugs to the assay plate. Fluorescent images taken using the Cytation high-content imaging platform (Bio-Tek Inc., VT, US). (D) MDA-MB-231 microspheres treated with 1 μM staurosporine for 72 h completely degraded. Bright-field images taken using the Cytation high-content imaging platform (Bio-Tek Inc., VT, US). (E) MDA-MB-231 microspheres treated with 10 μM doxorubicin are mostly intact but with visible cell damage. (F) Assessing the viability of MDA-MB-231 microspheres after 72 h of incubation using CellTiter-Glo 3D. Bright-field images taken using the Cytation high-content imaging platform (Bio-Tek Inc., VT, US). UC—untreated control, STS—staurosporine, DOX—doxorubicin (*p < 0.05, n = 5 wells per group. Scale bar = 200 μm).
A 384-well pin tool device (V&P Scientific, Inc. San Diego, CA) mounted on Biomek NXP (Beckman Coulter) was used for the administration of drug compounds. The diameter of a pin (0.457 mm) in the pin tool is slightly smaller than the size of our microspheres. Therefore, to determine whether compounds can be delivered into the wells containing microspheres without damaging or removing them, we pinned some doxorubicin and imaged wells before and after addition. When the number of microspheres was low (∼3), the microspheres were not perturbed by pinning (Figure 6C, top). When microspheres were tightly packed and pinning was done by touching pins to the bottom, pinning could potentially remove a microsphere from the well (Figure 6C, bottom). However, the shape of microspheres appeared to be unperturbed; therefore, we do not expect negative effects on viability. Alternatively, drugs can be delivered by pinning them into the media without touching the bottom of the well.
The MDA-MB-231 microspheres were also treated with drug compounds including 1 μM staurosporine and 10 μM doxorubicin in 384-well plates and incubated for 72 h. The microspheres treated with staurosporine were completely degraded (Figure 6D). The microspheres treated with doxorubicin were mostly intact, but encapsulated cells showed signs of cellular damage (Figure 6E). The degradation of the microspheres treated with staurosporine could be a result of the complete cancer cell degradation which releases multiple nonspecific proteinases capable of degrading the fibrinogen constituent of the microspheres.59 Doxorubicin-treated spheres are intact since cells are not completely degraded, and, as a result, the proteinases are not released. Quantitative analysis of viability showed that both the drug treatments were able to significantly reduce the cell viability compared to the untreated control group (Figure 6F).
For the drug-testing assays, all MDA-MB-231 microspheres were produced and then shipped overnight to a separate facility for drug testing. Demonstrating the ability to distribute the microspheres for third-party testing is an important step toward the establishment of ready-to-use tissue-engineered cancer models. In this way, end users can receive the microspheres, employ them for assays, and obtain desired information with minimal additional training. Having ready-to-use, shippable, spheroidal-engineered tissue cancer models would further streamline the automated drug testing process, leading to a more efficient drug discovery process. Taken together, these results suggest that the cancer microspheres have the potential to be used as models for HTS strategies.
Conclusions
In this study, we used a droplet-microfluidic system for rapid, facile, and reproducible fabrication of uniform cancer microspheres with high cell densities. The microfluidic system enables us to leverage tissue engineering tool sets in a spheroidal geometry to produce highly consistent engineered cancer tissues that can be directly employed in existing spheroidal cell aggregate assays. Fabricated hydrogel microspheres supported the 3D culture of breast cancer cells over at least 14 days in culture. Encapsulated cells displayed cell-type-specific differences in morphology, proliferation, metabolic activity, ultrastructure, and overall microsphere size distribution and bulk stiffness. The cancer microsphere platform was shown to be compatible with an automated liquid handling system for administration of drug compounds and displayed proper drug responses after the treatment, demonstrating initial applicability to HTS drug discovery. Providing more physiologically relevant drug response in a high-throughput and low-cost manner, the PF hydrogel-based cancer microspheres could potentially improve the translational success of drug candidates by providing more accurate in vitro prediction of in vivo drug efficacy. The developed cancer microsphere platform could enable further mechanistic investigations of cell–cell and cell–matrix interactions, tumorigenic phenomena, and disease progression. In addition, this microfluidic system could potentially be extended toward encapsulation of various other cell types for applications in tissue engineering and regenerative therapy, such as the production of cardiac or hepatic microspheres for injectable cell delivery or drug toxicity screening. Future work is needed to examine the utility of our cancer microspheres in drug screening against existing drug libraries, in comparison with 2D cell models, 3D self-aggregated cancer spheroids, and known in vivo response, to determine the ability to overcome limitations of current in vitro models in HTS.
Acknowledgments
This work was supported by the Auburn University Research Initiative in Cancer (AURIC) Graduate Fellowship (S.P.), AU-CMB/NSF EPSCoR NSF-EPS-1158862 summer research fellowship (W.J.S.), Alabama EPSCoR Graduate Research Scholarship Program (Y.T.), National Science Foundation NSF-CBET-1743445 (Y.T. and E.A.L.), and NIH grant R15CA249788 (D.M.). The authors are grateful to Dr. C. Ross Ethier, Wallace H. Coulter Department of Biomedical Engineering, Georgia Institute of Technology, Atlanta, GA, USA, for facilitating the use of the microsquisher system for mechanical characterization. The table of contents figure was created with BioRender.com.
Author Present Address
∥ Department of Biotechnology, Bhupat and Jyoti Mehta School of Biosciences, Indian Institute of Technology Madras, Chennai 600036, India
Author Contributions
⊥ W.J.S., Y.T., and S.P. contributed equally to this work.
The authors declare the following competing financial interest(s): Shantanu Pradhan is the technical advisor and co-director of ISMO Biophotonics Pvt. Ltd., registered in Chennai, India, and holds equity stake in this venture. Auburn University has one patent issued for the microfluidic cell encapsulation system employed. EA Lipke, Y Tian, WJ Seeto, Microfluidics Device for Fabrication of Large, Uniform, Injectable Hydrogel Microspheres for Cell Encapsulation. US Patent No. 11,166,920, issued 11/9/2021.
References
- Pradhan S.; Clary J. M.; Seliktar D.; Lipke E. A. A three-dimensional spheroidal cancer model based on PEG-fibrinogen hydrogel microspheres. Biomaterials 2017, 115, 141–154. 10.1016/j.biomaterials.2016.10.052. [DOI] [PubMed] [Google Scholar]
- Han S. J.; Kwon S.; Kim K. S. Challenges of applying multicellular tumor spheroids in preclinical phase. Cancer Cell Int. 2021, 21, 152. 10.1186/s12935-021-01853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta G.; Hsiao A. Y.; Ingram M.; Luker G. D.; Takayama S. Opportunities and challenges for use of tumor spheroids as models to test drug delivery and efficacy. J. Controlled Release 2012, 164, 192–204. 10.1016/j.jconrel.2012.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape J.; Emberton M.; Cheema U. 3D Cancer Models: The Need for a Complex Stroma, Compartmentalization and Stiffness. Front. Bioeng. Biotechnol. 2021, 9, 660502. 10.3389/fbioe.2021.660502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seliktar D. Designing cell-compatible hydrogels for biomedical applications. Science 2012, 336, 1124–1128. 10.1126/science.1214804. [DOI] [PubMed] [Google Scholar]
- Peppas N. A.; Hilt J. Z.; Khademhosseini A.; Langer R. Hydrogels in Biology and Medicine: From Molecular Principles to Bionanotechnology. Adv. Mater. 2006, 18, 1345–1360. 10.1002/adma.200501612. [DOI] [Google Scholar]
- Tibbitt M. W.; Anseth K. S. Hydrogels as extracellular matrix mimics for 3D cell culture. Biotechnol. Bioeng. 2009, 103, 655–663. 10.1002/bit.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice J. J.; Martino M. M.; De Laporte L.; Tortelli F.; Briquez P. S.; Hubbell J. A. Engineering the regenerative microenvironment with biomaterials. Adv. Healthcare Mater. 2013, 2, 57–71. 10.1002/adhm.201200197. [DOI] [PubMed] [Google Scholar]
- Bruns J.; Egan T.; Mercier P.; Zustiak S. P. Glioblastoma spheroid growth and chemotherapeutic responses in single and dual-stiffness hydrogels. Acta Biomater. 2022, 10.1016/j.actbio.2022.05.048. [DOI] [PubMed] [Google Scholar]
- Kim H. N.; Habbit N. L.; Su C. Y.; Choi N.; Ahn E. H.; Lipke E. A.; Kim D. H. Microphysiological Systems as Enabling Tools for Modeling Complexity in the Tumor Microenvironment and Accelerating Cancer Drug Development. Adv. Funct. Mater. 2019, 29, 1807553. 10.1002/adfm.201807553. [DOI] [Google Scholar]
- Singh A.; Brito I.; Lammerding J. Beyond Tissue Stiffness and Bioadhesivity: Advanced Biomaterials to Model Tumor Microenvironments and Drug Resistance. Trends Cancer 2018, 4, 281–291. 10.1016/j.trecan.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata E.; Sahai E. Tumor Microenvironment and Differential Responses to Therapy. Cold Spring Harbor Perspect. Med. 2017, 7, a026781. 10.1101/cshperspect.a026781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenhizer D.; Dean T.; D’Arcangelo E.; McGuigan A. P. The current landscape of 3D in vitro tumor models: what cancer hallmarks are accessible for drug discovery?. Adv. Healthcare Mater. 2018, 7, 1701174. 10.1002/adhm.201701174. [DOI] [PubMed] [Google Scholar]
- Santo V. E.; Rebelo S. P.; Estrada M. F.; Alves P. M.; Boghaert E.; Brito C. Drug screening in 3D in vitro tumor models: overcoming current pitfalls of efficacy read-outs. Biotechnol. J. 2017, 12, 1600505. 10.1002/biot.201600505. [DOI] [PubMed] [Google Scholar]
- Katt M. E.; Placone A. L.; Wong A. D.; Xu Z. S.; Searson P. C. In Vitro Tumor Models: Advantages, Disadvantages, Variables, and Selecting the Right Platform. Front. Bioeng. Biotechnol. 2016, 4, 12. 10.3389/fbioe.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K. M.; Lewis D.; Gerecht S. Bioinspired Hydrogels to Engineer Cancer Microenvironments. Annu. Rev. Biomed. Eng. 2017, 19, 109–133. 10.1146/annurev-bioeng-071516-044619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S.; Hassani I.; Clary J. M.; Lipke E. A. Polymeric Biomaterials for In Vitro Cancer Tissue Engineering and Drug Testing Applications. Tissue Eng., Part B 2016, 22, 470–484. 10.1089/ten.TEB.2015.0567. [DOI] [PubMed] [Google Scholar]
- Shang M.; Soon R. H.; Lim C. T.; Khoo B. L.; Han J. Microfluidic modelling of the tumor microenvironment for anti-cancer drug development. Lab Chip 2019, 19, 369–386. 10.1039/c8lc00970h. [DOI] [PubMed] [Google Scholar]
- Moroni L.; Boland T.; Burdick J. A.; De Maria C.; Derby B.; Forgacs G.; Groll J.; Li Q.; Malda J.; Mironov V. A.; et al. Biofabrication: A Guide to Technology and Terminology. Trends Biotechnol. 2018, 36, 384–402. 10.1016/j.tibtech.2017.10.015. [DOI] [PubMed] [Google Scholar]
- Chung B. G.; Lee K. H.; Khademhosseini A.; Lee S. H. Microfluidic fabrication of microengineered hydrogels and their application in tissue engineering. Lab Chip 2012, 12, 45–59. 10.1039/c1lc20859d. [DOI] [PubMed] [Google Scholar]
- Pradhan S.; Hassani I.; Seeto W. J.; Lipke E. A. PEG-fibrinogen hydrogels for three-dimensional breast cancer cell culture. J. Biomed. Mater. Res., Part A 2017, 105, 236–252. 10.1002/jbm.a.35899. [DOI] [PubMed] [Google Scholar]
- Ling Y.; Rubin J.; Deng Y.; Huang C.; Demirci U.; Karp J. M.; Khademhosseini A. A cell-laden microfluidic hydrogel. Lab Chip 2007, 7, 756–762. 10.1039/b615486g. [DOI] [PubMed] [Google Scholar]
- Tan W. H.; Takeuchi S. Monodisperse Alginate Hydrogel Microbeads for Cell Encapsulation. Adv. Mater. 2007, 19, 2696–2701. 10.1002/adma.200700433. [DOI] [Google Scholar]
- Kumachev A.; Greener J.; Tumarkin E.; Eiser E.; Zandstra P. W.; Kumacheva E. High-throughput generation of hydrogel microbeads with varying elasticity for cell encapsulation. Biomaterials 2011, 32, 1477–1483. 10.1016/j.biomaterials.2010.10.033. [DOI] [PubMed] [Google Scholar]
- Teh S. Y.; Lin R.; Hung L. H.; Lee A. P. Droplet microfluidics. Lab Chip 2008, 8, 198–220. 10.1039/b715524g. [DOI] [PubMed] [Google Scholar]
- Guo M. T.; Rotem A.; Heyman J. A.; Weitz D. A. Droplet microfluidics for high-throughput biological assays. Lab Chip 2012, 12, 2146–2155. 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- Yu L.; Chen M. C.; Cheung K. C. Droplet-based microfluidic system for multicellular tumor spheroid formation and anticancer drug testing. Lab Chip 2010, 10, 2424–2432. 10.1039/c004590j. [DOI] [PubMed] [Google Scholar]
- Guerzoni L. P. B.; Rose J. C.; Gehlen D. B.; Jans A.; Haraszti T.; Wessling M.; Kuehne A. J. C.; De Laporte L. Cell Encapsulation in Soft, Anisometric Poly(ethylene) Glycol Microgels Using a Novel Radical-Free Microfluidic System. Small 2019, 15, 1900692. 10.1002/smll.201900692. [DOI] [PubMed] [Google Scholar]
- Franco C. L.; Price J.; West J. L. Development and optimization of a dual-photoinitiator, emulsion-based technique for rapid generation of cell-laden hydrogel microspheres. Acta Biomater. 2011, 7, 3267–3276. 10.1016/j.actbio.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Lu Y. C.; Song W.; An D.; Kim B. J.; Schwartz R.; Wu M.; Ma M. Designing compartmentalized hydrogel microparticles for cell encapsulation and scalable 3D cell culture. J. Mater. Chem. B 2015, 3, 353–360. 10.1039/c4tb01735h. [DOI] [PubMed] [Google Scholar]
- Rao W.; Zhao S.; Yu J.; Lu X.; Zynger D. L.; He X. Enhanced enrichment of prostate cancer stem-like cells with miniaturized 3D culture in liquid core-hydrogel shell microcapsules. Biomaterials 2014, 35, 7762–7773. 10.1016/j.biomaterials.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira M. B.; Kossover O.; Mano J. F.; Seliktar D. Injectable PEGylated fibrinogen cell-laden microparticles made with a continuous solvent- and oil-free preparation method. Acta Biomater. 2015, 13, 78–87. 10.1016/j.actbio.2014.11.013. [DOI] [PubMed] [Google Scholar]
- Panda P.; Ali S.; Lo E.; Chung B. G.; Hatton T. A.; Khademhosseini A.; Doyle P. S. Stop-flow lithography to generate cell-laden microgel particles. Lab Chip 2008, 8, 1056–1061. 10.1039/b804234a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendukuri D.; Pregibon D. C.; Collins J.; Hatton T. A.; Doyle P. S. Continuous-flow lithography for high-throughput microparticle synthesis. Nat. Mater. 2006, 5, 365–369. 10.1038/nmat1617. [DOI] [PubMed] [Google Scholar]
- Pataky K.; Braschler T.; Negro A.; Renaud P.; Lutolf M. P.; Brugger J. Microdrop printing of hydrogel bioinks into 3D tissue-like geometries. Adv. Mater. 2012, 24, 391–396. 10.1002/adma.201102800. [DOI] [PubMed] [Google Scholar]
- Suzuki D.; Horigome K.; Kureha T.; Matsui S.; Watanabe T. Polymeric hydrogel microspheres: design, synthesis, characterization, assembly and applications. Polym. J. 2017, 49, 695–702. 10.1038/pj.2017.39. [DOI] [Google Scholar]
- Zhao Z.; Wang Z.; Li G.; Cai Z.; Wu J.; Wang L.; Deng L.; Cai M.; Cui W. Injectable Microfluidic Hydrogel Microspheres for Cell and Drug Delivery. Adv. Funct. Mater. 2021, 31, 2103339. 10.1002/adfm.202103339. [DOI] [Google Scholar]
- Sabhachandani P.; Sarkar S.; Mckenney S.; Ravi D.; Evens A. M.; Konry T. Microfluidic assembly of hydrogel-based immunogenic tumor spheroids for evaluation of anticancer therapies and biomarker release. J. Controlled Release 2019, 295, 21–30. 10.1016/j.jconrel.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.; Grist S. M.; Nasseri S. S.; Cheng E.; Hwang Y. C.; Ni C.; Cheung K. C. Core-shell hydrogel beads with extracellular matrix for tumor spheroid formation. Biomicrofluidics 2015, 9, 024118. 10.1063/1.4918754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M.; Choi J. W.; Ahrberg C. D.; Choi H. W.; Ha J. H.; Mun S. G.; Mo S. J.; Chung B. G. Generation of tumor spheroids using a droplet-based microfluidic device for photothermal therapy. Microsyst. Nanoeng. 2020, 6, 52. 10.1038/s41378-020-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriou P.; Li J.; Tornillo G.; McCloy T.; Barrow D. Droplet Microfluidics for Tumor Drug-Related Studies and Programmable Artificial Cells. Global Challenges 2021, 5, 2000123. 10.1002/gch2.202000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G.; Li F.; Zhao X.; Ma Y.; Li Y.; Lin M.; Jin G.; Lu T. J.; Genin G. M.; Xu F. Functional and Biomimetic Materials for Engineering of the Three-Dimensional Cell Microenvironment. Chem. Rev. 2017, 117, 12764–12850. 10.1021/acs.chemrev.7b00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L.; Mooney D. J. Biomaterials and emerging anticancer therapeutics: engineering the microenvironment. Nat. Rev. Cancer 2016, 16, 56–66. 10.1038/nrc.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y.; Lipke E. A. Microfluidic Production of Cell-Laden Microspheroidal Hydrogels with Different Geometric Shapes. ACS Biomater. Sci. Eng. 2020, 6, 6435–6444. 10.1021/acsbiomaterials.0c00980. [DOI] [PubMed] [Google Scholar]
- Seeto W. J.; Tian Y.; Pradhan S.; Kerscher P.; Lipke E. A. Rapid Production of Cell-Laden Microspheres Using a Flexible Microfluidic Encapsulation Platform. Small 2019, 15, 1902058. 10.1002/smll.201902058. [DOI] [PubMed] [Google Scholar]
- Pradhan S.; Smith A. M.; Garson C. J.; Hassani I.; Seeto W. J.; Pant K.; Arnold R. D.; Prabhakarpandian B.; Lipke E. A. A Microvascularized Tumor-mimetic Platform for Assessing Anti-cancer Drug Efficacy. Sci. Rep. 2018, 8, 3171. 10.1038/s41598-018-21075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S.; Chaudhury C. S.; Lipke E. A. Dual-phase, surface tension-based fabrication method for generation of tumor millibeads. Langmuir 2014, 30, 3817–3825. 10.1021/la500402m. [DOI] [PubMed] [Google Scholar]
- de Rutte J. M.; Koh J.; Di Carlo D. Scalable High-Throughput Production of Modular Microgels for In Situ Assembly of Microporous Tissue Scaffolds. Adv. Funct. Mater. 2019, 29, 1900071. 10.1002/adfm.201900071. [DOI] [Google Scholar]
- Mao A. S.; Shin J. W.; Utech S.; Wang H.; Uzun O.; Li W.; Cooper M.; Hu Y.; Zhang L.; Weitz D. A.; et al. Deterministic encapsulation of single cells in thin tunable microgels for niche modelling and therapeutic delivery. Nat. Mater. 2017, 16, 236–243. 10.1038/nmat4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienemann P. S.; Rossow T.; Mao A. S.; Vallmajo-Martin Q.; Ehrbar M.; Mooney D. J. Single cell-laden protease-sensitive microniches for long-term culture in 3D. Lab Chip 2017, 17, 727–737. 10.1039/c6lc01444e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikhi A.; de Rutte J.; Haghniaz R.; Akouissi O.; Sohrabi A.; Di Carlo D.; Khademhosseini A. Microfluidic-enabled bottom-up hydrogels from annealable naturally-derived protein microbeads. Biomaterials 2019, 192, 560–568. 10.1016/j.biomaterials.2018.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeto W. J.; Tian Y.; Winter R. L.; Caldwell F. J.; Wooldridge A. A.; Lipke E. A. Encapsulation of Equine Endothelial Colony Forming Cells in Highly Uniform, Injectable Hydrogel Microspheres for Local Cell Delivery. Tissue Eng., Part C 2017, 23, 815–825. 10.1089/ten.TEC.2017.0233. [DOI] [PubMed] [Google Scholar]
- McGuigan A. P.; Sefton M. V. Vascularized organoid engineered by modular assembly enables blood perfusion. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 11461–11466. 10.1073/pnas.0602740103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.; Kong T.; Yeung K. W.; Shum H. C.; Cheung K. M.; Wang L.; To M. K. Fabrication and characterization of monodisperse PLGA-alginate core-shell microspheres with monodisperse size and homogeneous shells for controlled drug release. Acta Biomater. 2013, 9, 7410–7419. 10.1016/j.actbio.2013.03.022. [DOI] [PubMed] [Google Scholar]
- Skop N. B.; Calderon F.; Levison S. W.; Gandhi C. D.; Cho C. H. Heparin crosslinked chitosan microspheres for the delivery of neural stem cells and growth factors for central nervous system repair. Acta Biomater. 2013, 9, 6834–6843. 10.1016/j.actbio.2013.02.043. [DOI] [PubMed] [Google Scholar]
- Impellitteri N. A.; Toepke M. W.; Lan Levengood S. K.; Murphy W. L. Specific VEGF sequestering and release using peptide-functionalized hydrogel microspheres. Biomaterials 2012, 33, 3475–3484. 10.1016/j.biomaterials.2012.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco D.; Tumarkin E.; Kumacheva E. Microfluidic encapsulation of cells in polymer microgels. Small 2012, 8, 1633–1642. 10.1002/smll.201102464. [DOI] [PubMed] [Google Scholar]
- Peela N.; Sam F. S.; Christenson W.; Truong D.; Watson A. W.; Mouneimne G.; Ros R.; Nikkhah M. A three dimensional micropatterned tumor model for breast cancer cell migration studies. Biomaterials 2016, 81, 72–83. 10.1016/j.biomaterials.2015.11.039. [DOI] [PubMed] [Google Scholar]
- Almany L.; Seliktar D. Biosynthetic hydrogel scaffolds made from fibrinogen and polyethylene glycol for 3D cell cultures. Biomaterials 2005, 26, 2467–2477. 10.1016/j.biomaterials.2004.06.047. [DOI] [PubMed] [Google Scholar]