Abstract
Phage AR1 is similar to phage T4 in several essential genes but differs in host range. AR1 infects various isolates of Escherichia coli O157:H7 but does not infect K-12 strains that are commonly infected by T4. We report here the determinants that confer this infection specificity. In T-even phages, gp37 and gp38 are components of the tail fiber that are critical for phage-host interaction. The counterparts in AR1 may be similarly important and, therefore, were characterized. The AR1 gp37 has a sequence that differs totally from those of T2 and T4, except for a short stretch at the N terminus. The gp38 sequence, however, has some conservation between AR1 and T2 but not between AR1 and T4. The sequences that are most closely related to the AR1 gp37 and gp38 are those of phage Ac3 in the T2 family. To identify the AR1-specific receptor, E. coli O157:H7 was mutated by Tn10 insertion and selected for an AR1-resistant phenotype. A mutant so obtained has an insertion occurring at ompC that encodes an outer membrane porin. To confirm the role of OmpC in the AR1 infection, homologous replacement was used to create an ompC disruption mutant (RM). When RM was complemented with OmpC originated from an O157:H7 strain, but not from K-12, its AR1 susceptibility was fully restored. Our results suggest that the host specificity of AR1 is mediated at least in part through the OmpC molecule.
K-12 strains of Escherichia coli are nonpathogenic and have been used extensively for biochemical and genetic research (4, 14). Pathogenic E. coli strains are relatively scarce, but they can cause various types of disease (35, 39). O157:H7 is the principal serotype of enterohemorrhagic E. coli (EHEC). It can cause watery diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome in humans (33). Some distinct features of EHEC have been defined. Similar to Shigella dysenteriae, EHEC produces cytotoxins that cause host cell death by inhibiting protein synthesis (27). Comparable to enteropathogenic E. coli, EHEC has genes clustered in the pathogenicity island that are important for the so-called attaching and effacing lesion (20, 21). Additional determinants that differentiate O157:H7 from other E. coli strains include the presence of serologically specific O and H antigens, the absence of beta-glucuronidase, the lack of biochemical enzymes for sorbitol fermentation (9, 15, 29), and the presence of an 8-kDa outer membrane protein that mediates the bacterial adherence to the INT407 cell and chicken ceca (46).
Genetic variations of O157:H7 strains could also be revealed by phage typing (1, 2). One coliphage, called AR1, infects E. coli O157:H7 with high specificity (34) but does not infect other serotypical strains, including K-12. The morphology and the nucleotide sequences coding for capsid proteins and alpha-glucosyltransferase of AR1 are similar to those of phage T4 (44). However, the SegD gene, which is nonessential to the T4 life cycle, is not conservatively observed in the AR1 genome. Moreover, T4 infects K-12 strains of E. coli, but AR1 does not cause the same infection. Reciprocally, T4 does not infect the AR1-susceptible O157:H7 strains. K-12 strain infection by T4 is through contact with OmpC (43), an outer membrane porin. Interestingly, OmpC of the O157:H7 strain varies from that of the K-12 strains (44). Whether this varied OmpC confers resistance to T4 infection remains unclear.
It is also known that T-even coliphages recognize their cellular receptors with the free ends of their six long tail fibers. In T4, the distal part of these fibers are trimeric, with a stoichiometry of gp34/gp37/gp36/gp35 of 3:3:3:1 (6). gp38, together with a second phage protein, gp57, catalyzes the organization of gp37 but is absent from the phage particle. gp37 is responsible for receptor recognition (24, 42). The situation is different for the otherwise very closely related phage T2. The C-terminal 130 residues are removed from the T2 gp37 during fiber assembly, and one copy of gp38, unrelated to that of T4, is bound to the tip of the tail fibers for receptor recognition (8).
Since AR1 has a peculiar host range, it may have specialized gp37 and gp38. Hence, we deduced the gene sequences and characterized the respective proteins. Our results suggest that the AR1 distal tail fiber is organized in a way similar to that of phage Ac3 in the T2 family. Furthermore, we used transposon mutagenesis and gene replacement to generate mutants that are no longer infected by AR1. Complementation experiments were carried out with these mutants, and the results suggested that OmpC contributes to the initial binding of AR1.
MATERIALS AND METHODS
Bacterial culture and phage preparation.
The bacteria, bacteriophages, and plasmids used in this study are listed in Table 1. Coliphage AR1 was propagated on E. coli O157:H7 strain 1266 as described previously (34, 44).
TABLE 1.
Bacterial strains, bacteriophages, and plasmids
| Strain, phage, or plasmid | Relevant feature(s) | Source or reference |
|---|---|---|
| Strains | ||
| JC1569 | F′::Tn10lac+Ts TcrmetB1 leu-6 his-1 arg-6 rec acY1 xyl-7 mtl-2 gal-6 str-104 tonA2 tsx-1λ− sup-E44 | J. Ou |
| HER1266′ | Mutant (Lac−) of O157:H7 (HER1266); AR1 sensitive | This study |
| TM | Tn10 mutant of HER1266′; AR1 resistant | This study |
| JM109 | F′ traD36 lacIq Δ(lacZ)M15 proA+B+/e14− (McrA−) Δ(lac-proAB)thi gyrA96 (nNalr) endA1 hsdR17(rK− mK+) relA1 supE44 recA1 | Stratagene |
| KM | Tn10 mutant of JM109; T4 resistant | This study |
| Bacteriophages | ||
| AR1 | O157:H7-specific coliphage | 34 |
| T4 | Coliphage | ATCC 11303-B4 |
| Plasmids | ||
| pBluescript II SK(+) | Cloning vector (Ampr) | Stratagene |
| pGex-2T | Expression vector (Ampr) | 38 |
| pGexO157OmpC | Derived from pGex-2T; expresses GST-OmpC(O) | This study |
| pTac-85 | Expression vector (Ampr) | 19 |
| pTacOmpC(O) | Derived from pTac-85; expresses OmpC(O) | This study |
| pTacOmpC(K) | Derived from pTac-85; expresses OmpC(K) | This study |
Phage DNA isolation.
Nucleic acids were extracted from a concentrated phage solution (1 ml) by adding 10 μg of RNase A (Boehringer Mannheim) and 100 U of DNase I (Boehringer Mannheim). After incubating at 37°C for 15 min, the mixture was adjusted to 1% sodium dodecyl sulfate (SDS) and digested with 20 U of proteinase K for 1 h. Thereafter, phenol and chloroform were added to the mixture to remove proteinaceous materials. The remaining nucleic acids were extracted according to standard procedures (36).
Protein expression.
The product translated from the g37 open reading frame (ORF) has a mass of about 121 kDa. To ensure its efficient expression in E. coli, gp37 was sectioned into fragments for protein expression. DNA (g37) was amplified with three different PCR primer pairs: p37N (CCGGATCCTAAAGGCGGTAGTATTGACG) and p37-2CR (CGGAGGAGTAGTTAAATCGGTTCCAT), p37M (ATGGATCCGATTTAACTACTCCTCCG) and p37-2R (TCAATGTATGCCGACTGCCCC), and p37M′ (CAGGATCCGATATTCTACTAATGGAACCG) and p37CR (TTACGGTACCCACGGTCCTGTTACTGCCA). The resulting fragments, cloned into pQE32 (Qiagen), separately encode the N-terminal one-third, the middle one-third, and the C-terminal two-thirds of gp37.
Due to the relatively small size of gp38, the entire g38 gene was PCR amplified with primers p38 (CCGGATCCATGGCAGTAACAGGACCGTGGG) and t60R (CGTTATCTTTGAACAAGCGAT), and the PCR product was ligated with SmaI-cut pQE32 to express recombinant gp38.
All these recombinant proteins contain a His tag prior to the N terminus. However, the expressed proteins formed inclusion bodies, and purification with a nickel column gave poor yields. Therefore, these fusion proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and recovered by electroelution for mouse polyclonal antiserum preparations.
Gel electrophoresis and immunoblotting.
The AR1 phage was purified by centrifugation through a 20% sucrose cushion as previously described (44). Proteins in an SDS sample buffer were separated by SDS-PAGE and visualized by staining with Coomassie brilliant blue. Alternatively, the separated proteins were directly transferred onto a nitrocellulose membrane. The bound proteins were then sequentially reacted with specific antisera, biotin-labeled secondary antibodies, and avidin-conjugated horseradish peroxidase. Finally, the membrane was developed with 4-chloro-1-naphthol and H2O2 as previously described (45).
Mutagenesis.
E. coli O157:H7 strain HER1266 was mutated with ethyl methanesulfonate (22) and screened for galactosidase-negative mutants whose colonies appeared white on MacConkey plates. One of these lac mutant isolates, named HER1266′, remained as susceptible to AR1 as the parental strain. It is referred to as the wild-type strain herein. HER1266′ and JC1569 were both grown at 30°C to an optical density at 600 nm (OD600) of 0.6, and 0.5-ml aliquots of the cultures were mixed for conjugation. After receiving an additional 1 ml of Luria broth (LB) the cells were incubated at 30°C for 3 h. The cells were then plated on M9 minimal agar plates and incubated at 30°C for 48 h. The resulting colonies were replicated on MacConkey agar plates supplemented with tetracycline (15 μg/ml) and grown at 30°C. Pink colonies were picked up and cultured in LB containing the same concentration of tetracycline at 42°C for the selection of mutants with Tn10 transposition. In a parallel experiment, E. coli JM109, a K-12 strain, was mutated by the same strategy but with the omission of the ethyl methanesulfonate mutagenesis.
Isolation of phage-resistant mutants.
To select strains resistant to AR1, a mixture of mutants was grown to an OD600 of 0.1 in LB at 37°C. A 1-ml culture was incubated with AR1 at a multiplicity of infection of 1 PFU/cell. After 15 min the culture was plated on LB plates containing 15 μg of tetracycline per ml. The outgrowing colonies were confirmed for the AR1-resistant phenotype by performing both liquid and plaque assays. A T4-resistant JM109 strain was obtained by a similar strategy.
Southern blotting.
Southern hybridization was performed by a standard procedure (36). DNA was digested with restriction enzymes according to the manufacturers' recommendations. A 1.3-kb DNA probe containing the IS10 region of Tn10 was generated by PCR amplification from the F′ plasmid of JC1569 with primers Sis10 (CTGATGAATCCCCTAATGATTTTG) and Ris10 (CTGAGAGATCCCCTCATAATTTCC). After confirmation of the PCR product, the DNA was purified with GeneClean III (Bio 101) and labeled with [α-32P]dCTP using the Rediprime II system (Amersham). A 1.1-kb ompC probe was prepared similarly, except that the DNA fragment was PCR amplified from HER1266 using primers Oc5end (AAAAGGATCCATGAAAGTTAAAGTACTGTCCC) and Oc3end (TTAGAACTGGTAAACCAGACCCAG).
Northern analysis.
The total bacterial RNA was prepared by using TRI reagent (Gibco Life). RNA was precipitated by centrifugation and washed with 75% ethanol. The isolated RNA was separated by electrophoresis in a 0.8% agarose gel and blotted onto nylon membranes according to the published procedure (36). Membranes were hybridized with the probes described above. Autoradiography analyses of both Southern and Northern blots were processed with a PhosphorImager (Molecular Dynamics).
Mapping the Tn10-mutated gene.
To determine the Tn10 insertion site, the IS10 probe-hybridized DNA was eluted and cloned into pBluescript II SK(+) (Stratagene). The cloned DNA fragment was confirmed by hybridization with the IS10 probe and then sequenced manually using the Sequenase 2.0 kit (U.S. Biochemicals).
Preparation of antiserum against OmpC of E. coli O157:H7.
The full-length ompC gene was amplified from chromosomal DNA of HER1266 with primers Oc5end and Oc3end. The PCR product was digested with BamHI and ligated into the BamHI/SmaI-digested pGex-2T (38). The resulting plasmid, pGexO157OmpC, encodes a full-length OmpC fused to the carboxyl terminus of glutathione S-transferase (GST). The GST-OmpC fusion protein was extracted from bacterial inclusion bodies and further purified by elution from SDS-polyacrylamide gels. The gel-purified recombinant protein was then used to raise mouse polyclonal antibodies.
OmpC expression from a plasmid.
DNA fragments containing the full-length ompC were PCR amplified separately from E. coli strains HER1266 and K-12 as described above. These PCR products were then digested with BamHI and inserted into BamHI/NruI-digested pTac-85 (19). The resulting plasmids, encoding the HER1266 and K-12 OmpC molecules, were designated pTacOmpC(O) and pTacOmpC(K), respectively. Expression of the OmpC proteins on these plasmids is under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible tac promoter at their respective initiation codons. For easy discussion, OmpC proteins derived from O157:H7 and K-12 are herein referred to as OmpC(O) and OmpC(K), respectively.
Mutating ompC by gene replacement.
The ompC gene of HER1266′ was mutated by the gene replacement method previously described (17), with a slight modification. The tetracycline-resistant (Tc) gene was excised from pBR322 with SspI/MscI digestion. It was then inserted into a unique NruI site within ompC that was previously cloned in pZero (44). ompC with the Tc gene insertion was then subcloned into SmaI/SalI-digested pKO3 (17), and the resulting plasmid was screened from JM109 transformants. The obtained plasmid was then electroporated into HER1266′. Cells were then recovered in 1 ml of SOC (2% Bacto Tryptone, 0.5% yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, and 20 mM glucose) for 1 h at 30°C before plating on LB agar containing chloramphenicol (10 μg/ml). Incubation was continued at 30°C overnight. A colony was inoculated into 3 ml of chloramphenicol-containing LB and cultured at 30°C until the OD600 reached 0.6. The cells were then plated on prewarmed chloramphenicol-containing LB plates and incubated at 43°C. From these plates, five colonies were picked up and inoculated into 1 ml of LB. After incubation at 30°C for 5 h, the cells were plated at 30°C on 5% (wt/vol) sucrose plates. The outgrowing bacteria were plated in replicates on plates with or without chloramphenicol at 30°C. Replacement mutants (RM) that appeared chloramphenicol sensitive due to a loss of the replacement vector were selected.
Phage binding assay.
HER1266′ was starved in Met- and Cys-deficient RPMI medium for 1 h and then washed twice with the same medium. The bacterial pellet was then resuspended in the same medium and adjusted to an OD600 of 1.0. A total of 0.1 ml of AR1 stock and 18 μl of [35S]Met/Cys (14.5 μCi per μl) were added to 0.3 ml of the bacterial solution. After 5 h of incubation AR1 was precipitated from the supernatant by polyethylene glycol and purified by centrifugation through a sucrose cushion as described previously (44). This isotope-labeled AR1 was resuspended in 200 μl of phosphate-buffered saline (PBS).
To assay phage binding, appropriate amounts of phage and bacteria were mixed in duplicates and incubated at 4°C for 2 h. The bacteria were then pelleted in a microcentrifuge and washed twice with cold PBS. The relative amount of AR1 bound on the bacteria was monitored by a liquid scintillation counter (Wallac). The relative binding efficiency was calculated by referring to those of HER1266′ and the ompC RM as 100 and 0%, respectively.
Nucleotide sequence accession number.
The deduced AR1 DNA sequence encompassing the 3′-end regions of g36, g37, and g38 and the 5′-end majority of t is available from GenBank under accession no. AF208841.
RESULTS
Putative g37 and g38 of AR1.
For T-even phages, there are two completely different organizations of the distal tail fiber locus among all the T4-like phages: the T2 and the T4 forms. gp37 is present in the phage particles of both forms (11, 24), while gp38 can be found only in the mature phage particle of the T2 form (25, 32). There are three families that have the tail fiber organization of the T2 form: T2-like, T6-like, and Ac3-like sequences (40). The sequences of g37 and g38 vary to different degrees among these phages, whereas those of g36 and t flanking this locus are quite conserved. This property permits facile amplification by PCR and sequencing analysis. Thus, we amplified and sequenced a DNA segment covering the 3′ ends of g36, g37, and g38 and the 5′ end of t. As expected, the g36 and t fragments were found to be highly homologous to the T4 counterparts. The putative g37 of AR1 has a 3,309-bp ORF that theoretically encodes a 1,103-residue protein. The size of this protein is between that of T4 (1,026 residues) and that of T2 (1,341 residues). Except for the N-terminal region, the AR1 gp37 totally differs from the gp37 of either T4 or T2 (data not shown). It is believed that gp37 interacts with gp36 through its N-terminal domain (3, 31). Therefore, it is not surprising that the N-terminal 50 residues are nearly identical in the known T-even and related phages (40).
The N-terminal homology confirms the assignment of this 3,309-bp ORF as g37 of AR1. However, additional stretches of sequences that are homologous between gp37 of T4 and that of T2 are not observed in similar paired comparisons involving AR1 (data not shown). The AR1 gp37 has an additional stretch spanning 136 residues near the C terminus that are homologous (61% identity) to those of T6. More closely related, the AR1 gp37 has the same length as and a high degree (76%) of amino acid identity to that of Ac3.
The putative gp38 of AR1 has an ORF encoding 259 amino acids. The size of the protein is closer to that of T2 (262 residues) than to that of T4 (183 residues). The best alignment with the AR1 gp38 sequence is obtained with that of phage Ac3, which gives 69% identity in the entire sequence. Highly conserved regions are observed in the N-terminal half, the C-terminal 22 residues, and characteristic oligoglycine stretches of the protein. The conserved glycine-rich stretches have been suggested to form omega loops that position the adhesin sequence on the exterior of gp38 (16). Therefore, the sequence comparison results from g37 and g38 suggest that AR1 has a tail fiber organization similar to that of Ac3. It is therefore likely that the AR1 gp38 may be present in the mature phage particle and contribute to its specific binding to E. coli O157:H7.
Characterization of gp37 and gp38.
To confirm the presence of the putative g37 and g38 products in the AR1 phage particles, several mouse antisera were generated with recombinant proteins as immunogens. Three recombinant gp37 fragments that separately contain the N-terminal portion, the middle part, and the C-terminal two-thirds of gp37 were generated and used for animal immunization. The obtained antisera were then used to react with proteins prepared from the AR1 particles by Western blotting. A 100-kDa protein, slightly smaller than the one predicted from the ORF, was detected by all three antisera (data not shown). This result suggests that either the intact AR1 gp37 moves electrophoretically faster than expected or the protein may undergo a C-terminal maturation processing during the tail fiber assembly, as in the case of T2 (8). To ensure that the product of g38 is present in the phage particle, the entire g38 ORF was expressed as a recombinant protein. The recombinant gp38-generated antiserum detected a 27-kDa protein in the phage lysate (data not shown) in an analysis similar to that described above. The size of this protein is consistent with that predicted from the g38 ORF. Therefore, these protein data were consistent with the results deduced from DNA sequencing, supporting the idea that AR1 has an Ac3-like tail fiber structure.
AR1-resistant mutants obtained by transposon mutagenesis.
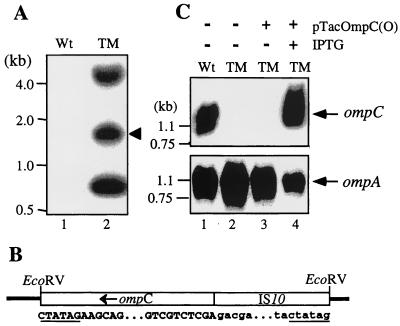
One of the AR1-resistant O157:H7 mutants, named TM, was selected for further examination. To ensure that Tn10 had been inserted into the chromosome of TM, bacterial DNA was extracted and digested with EcoRV, which is known to make a total of three cuts in the IS10 sequences of Tn10: two cuts in IS10L and one in the slightly different IS10R. Results from Southern analysis (Fig. 1A) show that unlike the wild-type control (lane 1), TM (lane 2) has three DNA fragments hybridized with the IS10 probe, a fact reflecting the nature of Tn10 mutagenesis.
FIG. 1.
Analysis of an E. coli O157:H7 mutant (TM) created by Tn10 transposition. (A) Southern blot analysis of bacterial chromosomal DNAs that had been digested with EcoRV and hybridized with an IS10 probe. (B) Schematic diagram of the sequenced IS10-containing fragment in TM. The EcoRV fragment of TM (indicated by an arrowhead in panel A) was inserted into EcoRV-cut pBluescript II SK(+) and sequenced. The junctions between the insert (open box) and the vector (thick line) are illustrated, and portions of the cloned sequence are shown beneath. The EcoRV restriction sites are underlined. The sequence in the middle portion illustrates where ompC (capital letters) was disrupted by the IS10 insertion (lowercase letters). (C) Northern blot analysis of RNAs isolated from the TM mutant. Total bacterial RNAs were extracted from different preparations of bacteria. The RNA (10 μg) was electrophoretically separated on an agarose gel, transferred to a nylon membrane, and hybridized with ompC (upper panel) and ompA (lower panel) probes, respectively. In this and the following experiments (Fig. 2), gene expression from the plasmids was induced with IPTG (1 mM). Wt, wild-type strain.
The 0.66-kb fragment was presumably the EcoRV fragment of IS10L, whereas the 1.8-kb and the >4.0-kb fragments might represent the EcoRV fragments containing a portion of IS10 and a flanking sequence. To explore this possibility, the 1.8-kb EcoRV fragment was cloned and sequenced from both ends, and the results are summarized in Fig. 1B. This cloned EcoRV fragment comprises 939 bp from IS10 and 910 bp from ompC. The gene ompC was apparently disrupted at the 5′ end, since IS10 was placed directly upstream of the 33rd nucleotide of ompC. Southern blot analysis using a probe derived from the full-length ompC (data not shown) further confirmed that ompC in TM was disrupted.
OmpC is an outer membrane porin and serves as a receptor for bacteriophages TuIb and T4 (24). It may have a similar function for AR1. Therefore, we first examined the OmpC expression in TM at both the RNA and protein levels. Then we constructed tac promoter-driven expression vectors pTacOmpC(O) and pTacOmpC(K) to express OmpC of O157:H7 and K-12 origins (44), respectively. These plasmids were thereafter used in complementation assays to address whether the loss of the AR1 susceptibility could be restored by a plasmid-encoded OmpC.
The expression of ompC-specific mRNAs in bacteria was analyzed with Northern blotting (Fig. 1C, upper panel). The ompC probe did not detect any signal from the RNA sample of TM (lane 2), while a strong signal was observed for a 1.3-kb RNA isolated from the wild-type control (lane 1). RNA extracted from TM transformed with pTacOmpC(O) was also included in the analysis. In the presence of IPTG, ompC-specific RNA was drastically induced (lane 4) compared to that without the IPTG addition (lane 3). The absence of ompC-specific RNA in lanes 2 and 3 did not result from poor RNA preparations, since the same samples could hybridize to a probe of ompA (Fig. 1C, lower panel), which encodes a protein (OmpA) functioning as a receptor for phages K3, Ox2, and M1 (10, 18, 26).
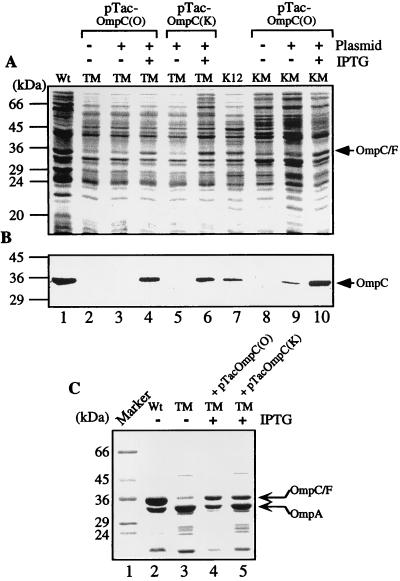
The total bacterial proteins were then analyzed by SDS-PAGE separation and visualized by Coomassie blue dye staining. Figure 2A shows a 33-kDa protein band that decreased in intensity in TM (lane 2) compared to that in the wild-type strain (lane 1). The intensity of this 33-kDa band was restored after TM was transformed with pTacOmpC(O) and cultured in the presence of IPTG (lane 4). The amount of the 33-kDa protein did not increase significantly when IPTG was omitted from the culture medium (lane 3). Similarly, expression of the 33-kDa protein in TM was restored upon receiving pTacOmpC(K), which encodes OmpC of the K-12 origin (compare lanes 5 and 6).
FIG. 2.
Protein expression patterns of the wild-type and mutant E. coli strains. (A) Coomassie blue-stained profiles of the total proteins separated on an SDS–12% polyacrylamide gel. (B) Western blot analysis of OmpC expressed in the total bacterial lysates using mouse anti-OmpC antisera. (C) Coomassie blue-stained protein profiles of the outer membrane fractions. The identity of OmpA was confirmed by N-terminal microsequencing. Plasmids pTacOmp(O) and pTacOmp(K) encode OmpC of the O157:H7 origin and that of the K-12 origin, respectively. OmpC/F labels the band containing both OmpC and OmpF that are presumably not separated in these gel systems. Wt, wild-type strain.
OmpF has been known to comigrate with OmpC on SDS-PAGE gels (5, 37). We performed Western blotting analysis to ascertain that the changes in cellular OmpC concentration can be reflected by the fluctuating intensity of the 33-kDa protein band. OmpC of E. coli O157:H7 and that of the K-12 strain share 93.5% amino acid identity (44). Polyclonal antibodies raised against OmpC of E. coli O157:H7 would react with that of both origins (Fig. 2B). Indeed, the antigen (OmpC) was present abundantly in the wild-type strain (lane 1) but was absent from the mutant strain (lane 2). In the presence of IPTG, both pTacOmpC(O)- and pTacOmpC(K)-transformed TM mutants expressed OmpC to a high level (lanes 4 and 6). In the absence of IPTG, OmpC was not expressed efficiently (lanes 3 and 5). A small amount of OmpC could still be detected when the blot was developed longer, a result presumably due to the leakage of the tac promoter (also see Fig. 2B, lane 9).
To examine whether the plasmid-expressed OmpC in TM could be transported to the membrane as in the wild-type control, the bacterial membrane fractions were analyzed. Figure 2C shows the membrane protein patterns of a Coomassie blue dye-stained SDS-PAGE gel. The intensity of the OmpC/OmpF band was heavier than that of OmpA, a porin deduced by the N-terminal microsequencing, in the wild-type strain (lane 2), whereas OmpA was the dominant band in TM (lane 3). Presumably, the residual 33-kDa band detected in TM represented the OmpF molecules. The TM transformants with pTacOmpC(O) (lane 4) or pTacOmpC(K) (lane 5) expressed OmpC and had a substantial increase in the 33-kDa band intensity. However, as determined by comparison to the amount of OmpA in the same preparation, the amount of OmpC in the outer membrane of the plasmid-transformed strains had not been fully restored to the level of the wild-type control.
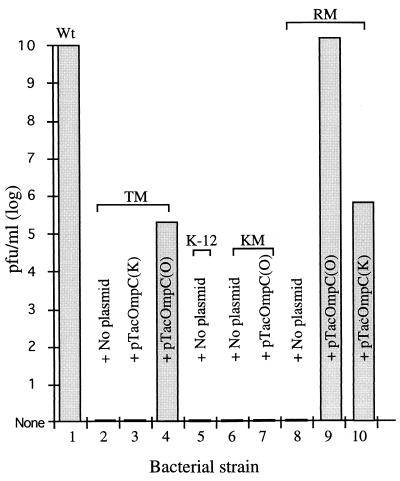
Whether the plasmid-expressed OmpC can restore the AR1 susceptibility of TM was then addressed. As shown in Fig. 3, a stock of phage that gave a titer of 1010 PFU/ml on the wild-type strain (column 1) gave no PFU on TM (column 2). The infection titer rose to 2 × 105 PFU/ml after TM was transformed with pTacOmpC(O) and cultured in the presence of IPTG (column 4). This recovery in plating efficiency did not occur with a similarly treated transformant harboring pTacOmpC(K) (column 3). These facts indicate that OmpC of the O157:H7 origin is essential for AR1 infection. However, providing this molecule to TM is not sufficient to recover all the lost AR1 plating efficiency.
FIG. 3.
AR1 plaque formation on different preparations of bacteria. The AR1 phage stock was made on the wild-type O157:H7 strain and diluted in a 10-fold series. The phage solutions were then used to infect various preparations of bacteria and plated out on LB agar plates for PFU scoring. When transformed with plasmids, the bacteria were cultivated in the presence of IPTG (1 mM) prior to mixing with the phage. IPTG (1 mM) was also included in the plates used in the plaque assay. No plaque formation was observed with bacteria indicated in columns 2, 3, and 5 through 8. These experiments were repeated three times, with similar results. Wt, wild-type strain.
E. coli K-12 strains are not susceptible to AR1 (44) (Fig. 3, column 5), and apparently this is due to the differences in OmpC (column 3). Whether introducing OmpC of the O157:H7 origin into a K-12 strain can overcome the host barrier to AR1 was examined. A JM109 mutant named KM that was generated similarly to TM was used in these experiments. Upon SDS-PAGE and Western blotting analyses (Fig. 2A and B, column 7 and 8), the 33-kDa protein band was diminished in KM compared to that of the parental K-12 strain. KM transformed with pTacOmpC(O) and cultivated in the presence of IPTG revived the expression of OmpC, as evidenced by Coomassie blue staining (Fig. 2A, columns 8 through 10) and Western blotting (Fig. 2B, columns 8 through 10). However, no plaques were produced when AR1 was applied to the pTacOmpC(O)-transformed KM (Fig. 3, column 7). These results indicated that another factor(s) besides OmpC contributes to the noninfectivity of the K-12 strains.
AR1-resistant mutants created by gene replacement.
To precisely mutate bacterial ompC, we generated a homologous RM of the O157:H7 strain by replacing the wild-type ompC with a version that had an insertion of a Tc gene sequence (17). This mutant was then confirmed by analyzing the ompC fragments by both PCR amplification and Southern blotting. Also as expected, no expression of OmpC in RM was observed (data not shown).
RM was then examined for susceptibility to AR1 as described above. A stock of AR1 containing 1010 PFU per ml on the wild-type O157:H7 gave no plaque on RM (Fig. 3, column 8). The AR1 plating efficiency was completely restored and gave 1.3 × 1010 PFU per ml when RM was transformed with pTacOmpC(O) and cultivated in the presence of IPTG (column 9). Interestingly, RM transformed with pTacOmpC(K) and cultivated similarly had 6.18 × 105 PFU/ml (column 10). This result was noticeably different from that observed with TM transformed with the same pTacOmpC(K) plasmid (compare columns 3 and 10) and supports the notion that TM had more genes affected than RM did when these mutations were created.
Binding of AR1 to different bacteria.
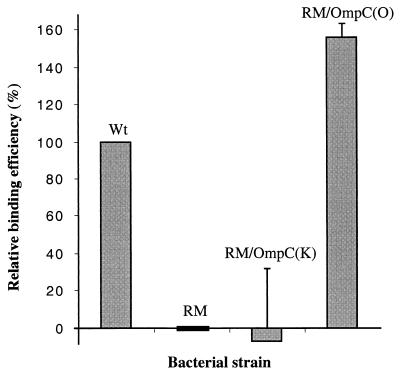
To demonstrate the receptor function of OmpC, an AR1 absorption assay was performed at 4°C to allow phage binding but not penetration. AR1 was labeled with [35S]Met and [35S]Cys and then was absorbed to different hosts. After incubation and washing, radioactivity of the phage associating with bacteria was counted and compared with that absorbed to the wild-type (as 100%) and the RM (as 0%) strains. Figure 4 shows that RM complemented with OmpC(O) had a slightly higher binding efficiency (1.5-fold) than the wild-type strain. This result is consistent with the 1.3-fold increase in the plaque-forming-efficiency assay (Fig. 3, columns 1 and 9). On the other hand, the RM transformant expressing OmpC(K) had a low binding activity indistinguishable from RM. Therefore, this system is unable to detect the weak phage-host interaction contributed from OmpC(K) expressed on RM.
FIG. 4.
AR1 binding assay. 35S-labeled AR1 phages were incubated at 4°C for 2 h with different bacteria as indicated. Cells were collected by centrifugation and washed twice with cold PBS. Bacterium-bound AR1 was then quantitated by liquid scintillation counting. Counts derived from the wild-type strain and RM were set as reference points for 100 and 0%, respectively.
DISCUSSION
AR1 is similar to the T-even phages in morphology, capsid genes, and genes encoding essential enzymes (44). However, AR1 has a unique host specificity that might be attributed to its distal tail fiber gp37 and gp38 proteins. We took advantage of the conserved g36 and t for PCR amplification and analyzed the g37 and g38 sequences between g36 and t. The N-terminal region of gp37 responsible for interacting with gp36 is structurally conserved with those of the known T-even phages (28, 30). Aside from this region, the structure of the AR1 gp37 diversifies distantly from those of T2 and T4 and is related to Ac3 and T6. This evolutionary relationship of the distal tail fiber could be similarly reflected by the gp38 of these phages: AR1 is homologous to Ac3, T6, and T2, in decreasing order, and is distant from T4.
The host receptor that interacts with AR1 appears to be OmpC of the O157:H7 origin. However, additional components in O157:H7 strains may cooperatively determine the phage infection efficiency. In TM, the loss of OmpC could be restored by transformation with either pTacOmp(O) or pTacOmp(K). However, only transformation with pTacOmpC(O) restored the sensitivity of TM to AR1. Therefore, OmpC molecules of the right identity and, perhaps, conformation appeared to be more important than the amount of OmpC present. Indeed, OmpC of the O157:H7 origin differs from that of K-12 by amino acid substitutions at 15 positions, a five-residue deletion, and a four-residue insertion in its total of 366 amino acids (44). A major varied segment is located between residues 176 and 226 where changes have been reported to affect the infectivity of several phages that use OmpC as the receptor (13, 23, 41). We speculate that the variations within this region of OmpC may affect, in large measure, the interaction with AR1. This notion has been further supported by the data from spontaneous mutants of E. coli O157:H7 that are resistant to AR1. Two of these mutants were analyzed for their ompC gene by PCR and direct sequencing (data not shown). The two isolates have the same mutations in OmpC: one Gly insertion between residues Ser178 and Glu179 and a Phe-to-Val alteration at the C-terminal residue 363.
In the pTacOmpC(O)-transformed TM, the recovered AR1 plating efficiency is significantly increased compared to that of TM. Nonetheless, this efficiency represented only a small fraction of that in the wild-type strain. We noticed that OmpC was not as efficiently translocated into the outer membrane as the wild-type control (Fig. 2C). The amount of OmpC on the membrane decreased severalfold, but this could not explain a 5-log loss of plating efficiency. AR1 plating efficiency was completely restored by transforming RM with the same plasmid. This observation rules out the possibility that the quantity of OmpC on the membrane is a detrimental factor to infection with the phage. We reasoned that TM, generated by Tn10 insertion, might have an additional gene(s) other than ompC affected by the transposon mutagenesis. Our preliminary results with TM have suggested that there is an additional Tn10 inserted in the ORF of waaJ, a gene that is involved in the biosynthesis of lipopolysaccharide (LPS) (12). In the absence of intact LPS, OmpC(O) presented on the bacterial surface may possibly have a low binding affinity for AR1. A similar small degree of AR1 plating efficiency was observed with RM presenting OmpC(K) (Fig. 3, lane 10). This result also suggests that a weak AR1-host interaction occurs when OmpC(K) is presented together with LPS from an O157:H7 strain.
To yield a maximum AR1 plating efficiency, the bacteria may require a cooperative interaction between OmpC and LPS, and both OmpC and LPS have to originate from the O157:H7 strain. OmpC(O) functions as the phage binding receptor, whereas the role of O157:H7-specific LPS remains unclear. One possibility is that O157:H7-specific LPS acts directly to enhance the phage binding affinity. Alternatively, it may function indirectly in assisting outer membrane biogenesis and folding of the membrane proteins (7). Undoubtedly, more experiments are needed to clarify this issue. Nonetheless, our present data demonstrate that AR1 has a variant of the distal tail fiber locus and that the host bacteria (O157:H7 strains) have a unique OmpC to serve as the phage receptor. These lines of information provide an answer to why AR1 specifically infects a serotype of E. coli (34, 44) that is associated with hemorrhagic diarrhea.
ACKNOWLEDGMENTS
We thank G. M. Church, J. T. Ou, and S. T. Hu for providing valuable reagents. We also thank M. Tam for critical reading of the manuscript.
S.-L. Yu and K.-L. Ko contributed equally to this work.
This research was financially supported in part by grant DOH88-HR-606 from the National Health Research Institute, grant NSC89-2320-B010-025 from the National Science Council, and an award from the Medical Research Advancement Foundation in memory of Chi-Shuen Tsou.
REFERENCES
- 1.Ahmed R, Bopp C, Borczyk A, Kasatiya S. Phage-typing scheme for Escherichia coli O157:H7. J Infect Dis. 1987;155:806–809. doi: 10.1093/infdis/155.4.806. [DOI] [PubMed] [Google Scholar]
- 2.Barrett T J, Lior H, Green J H, Khakhria R, Wells J G, Bell B P, Greene K D, Lewis J, Griffin P M. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J Clin Microbiol. 1994;32:3013–3017. doi: 10.1128/jcm.32.12.3013-3017.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beckendorf S K. Structure of the distal half of the bacteriophage T4 tail fiber. J Mol Biol. 1973;73:37–53. doi: 10.1016/0022-2836(73)90157-5. [DOI] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Braun-Breton C, Hofnung M. In vivo and in vitro functional alterations of the bacteriophage lambda receptor in lamB missense mutants of Escherichia coli K-12. J Bacteriol. 1981;148:845–852. doi: 10.1128/jb.148.3.845-852.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerritelli M E, Wall J S, Simon M N, Conway J F, Steven A C. Stoichiometry and domainal organization of the long tail-fiber of bacteriophage T4: a hinged viral adhesin. J Mol Biol. 1996;260:767–780. doi: 10.1006/jmbi.1996.0436. [DOI] [PubMed] [Google Scholar]
- 7.de Cock H, Brandenburg K, Wiese A, Holst O, Seydel U. Non-lamellar structure and negative charges of lipopolysaccharides required for efficient folding of outer membrane protein PhoE of Escherichia coli. J Biol Chem. 1999;274:5114–5119. doi: 10.1074/jbc.274.8.5114. [DOI] [PubMed] [Google Scholar]
- 8.Drexler K, Riede I, Henning U. Morphogenesis of the long tail fibers of bacteriophage T2 involves proteolytic processing of the polypeptide (gene product 37) constituting the distal part of the fiber. J Mol Biol. 1986;191:267–272. doi: 10.1016/0022-2836(86)90263-9. [DOI] [PubMed] [Google Scholar]
- 9.Farmer J J, III, Davis B R. H7 antiserum-sorbitol fermentation medium: a single tube screening medium for detecting Escherichia coli O157:H7 associated with hemorrhagic colitis. J Clin Microbiol. 1985;22:620–625. doi: 10.1128/jcm.22.4.620-625.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock R E W, Reeves P. Bacteriophage resistance in Escherichia coli K-12: general pattern of resistance. J Bacteriol. 1975;121:983–993. doi: 10.1128/jb.121.3.983-993.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashemolhosseini S, Holmes Z, Mutschler B, Henning U. Alterations of receptor specificities of coliphages of the T2 family. J Mol Biol. 1994;240:105–110. doi: 10.1006/jmbi.1994.1424. [DOI] [PubMed] [Google Scholar]
- 12.Heinrichs D E, Yethon J A, Whitfield C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol Microbiol. 1998;30:221–232. doi: 10.1046/j.1365-2958.1998.01063.x. [DOI] [PubMed] [Google Scholar]
- 13.Hikita C, Satake Y, Yamada H, Mizuno T, Mizushima S. Structural and functional characterization of the OmpF and OmpC porins of the Escherichia coli outer membrane: studies involving chimeric proteins. Res Microbiol. 1989;140:177–190. doi: 10.1016/0923-2508(89)90074-0. [DOI] [PubMed] [Google Scholar]
- 14.Itoh T, Aiba H, Baba T, Fujita K, Hayashi K, Inada T, Isono K, Kasai H, Kimura S, Kitakawa M, Kitagawa M, Makino K, Miki T, Mizobuchi K, Mori H, Mori T, Motomura K, Nakade S, Nakamura Y, Nashimoto H, Nishio Y, Oshima T, Saito N, Sampei G, Seki Y, Sivasundaram S, Tagami H, Takeda J, Takemoto K, Wada C, Yamamoto Y, Horiuchi T. A 460-kb DNA sequence of the Escherichia coli K-12 genome corresponding to the 40.1-50.0 min region on the linkage map. DNA Res. 1996;3:379–392. doi: 10.1093/dnares/3.6.379. [DOI] [PubMed] [Google Scholar]
- 15.Kleanthous H, Fry N K, Smith H R, Gross R J, Rowe B. The use of sorbitol-MacConkey agar in conjunction with a specific antiserum for the detection of Vero cytotoxin-producing strains of Escherichia coli O157. Epidemiol Infect. 1988;101:327–335. doi: 10.1017/s0950268800054261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leszczynski J F, Rose G D. Loops in globular proteins: a novel category of secondary structure. Science. 1986;234:849–855. doi: 10.1126/science.3775366. [DOI] [PubMed] [Google Scholar]
- 17.Link A J, Phillips D, Church G M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manning P A, Puspurs A, Reeves P. Outer membrane of Escherichia coli K-12: isolation of mutants with altered protein 3A by using host range mutants of bacteriophage K3. J Bacteriol. 1976;127:1080–1084. doi: 10.1128/jb.127.3.1080-1084.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh P. Ptac-85, an E. coli vector for expression of non-fusion proteins. Nucleic Acids Res. 1986;14:3603. doi: 10.1093/nar/14.8.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McDaniel T K, Kaper J B. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 22.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 23.Mizuno T, Kasai H, Mizushima S. Construction of a series of ompC-ompF chimeric genes by in vivo homologous recombination in Escherichia coli and characterization of their translational products. Mol Gen Genet. 1987;207:217–223. doi: 10.1007/BF00331581. [DOI] [PubMed] [Google Scholar]
- 24.Montag D, Hashemolhosseini S, Henning U. Receptor-recognizing proteins of T-even type bacteriophages. The receptor-recognizing area of proteins 37 of phages T4 TuIa and TuIb. J Mol Biol. 1990;216:327–334. doi: 10.1016/S0022-2836(05)80324-9. [DOI] [PubMed] [Google Scholar]
- 25.Montag D, Riede I, Eschbach M L, Degen M, Henning U. Receptor-recognizing proteins of T-even type bacteriophages. Constant and hypervariable regions and an unusual case of evolution. J Mol Biol. 1987;196:165–174. doi: 10.1016/0022-2836(87)90519-5. [DOI] [PubMed] [Google Scholar]
- 26.Morona R, Klose M, Henning U. Escherichia coli K-12 outer membrane protein (OmpA) as a bacteriophage receptor: analysis of mutant genes expressing altered proteins. J Bacteriol. 1984;159:570–578. doi: 10.1128/jb.159.2.570-578.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Brien A D, Newland J W, Miller S F, Holmes R K, Smith H W, Formal S B. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science. 1984;226:694–696. doi: 10.1126/science.6387911. [DOI] [PubMed] [Google Scholar]
- 28.Oliver D B, Crowther R A. DNA sequence of the tail fibre genes 36 and 37 of bacteriophage T4. J Mol Biol. 1981;153:545–568. doi: 10.1016/0022-2836(81)90407-1. [DOI] [PubMed] [Google Scholar]
- 29.Padhye N V, Doyle M P. Escherichia coli O157:H7: epidemiology, pathogenesis, and methods for detection in food. J Food Prot. 1992;55:555–565. doi: 10.4315/0362-028X-55.7.555. [DOI] [PubMed] [Google Scholar]
- 30.Riede I, Drexler K, Eschbach M L. The nucleotide sequences of the tail fiber gene 36 of bacteriophage T2 and of genes 36 of the T-even type Escherichia coli phages K3 and Ox2. Nucleic Acids Res. 1985;13:605–616. doi: 10.1093/nar/13.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riede I, Drexler K, Eschbach M L, Henning U. DNA sequence of genes 38 encoding a receptor-recognizing protein of bacteriophages T2, K3 and of K3 host range mutants. J Mol Biol. 1987;194:31–39. doi: 10.1016/0022-2836(87)90713-3. [DOI] [PubMed] [Google Scholar]
- 32.Riede I, Drexler K, Schwarz H, Henning U. T-even-type bacteriophages use an adhesin for recognition of cellular receptors. J Mol Biol. 1987;194:23–30. doi: 10.1016/0022-2836(87)90712-1. [DOI] [PubMed] [Google Scholar]
- 33.Riley L W. The epidemiologic, clinical, and microbiologic features of hemorrhagic colitis. Annu Rev Microbiol. 1987;41:383–407. doi: 10.1146/annurev.mi.41.100187.002123. [DOI] [PubMed] [Google Scholar]
- 34.Ronner A B, Cliver D O. Isolation and characterization of a coliphage specific for Escherichia coli O157:H7. J Food Prot. 1990;53:944–947. doi: 10.4315/0362-028X-53.11.944. [DOI] [PubMed] [Google Scholar]
- 35.Salyers A A, Whitt D D. Bacterial pathogenesis: a molecular approach. Washington, D.C.: ASM Press; 1994. [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Schneider H, Fsihi H, Kottwitz B, Mygind B, Bremer E. Identification of a segment of the Escherichia coli Tsx protein that functions as a bacteriophage receptor area. J Bacteriol. 1993;175:2809–2817. doi: 10.1128/jb.175.10.2809-2817.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in E. coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 39.Su C, Lawrence J B. Escherichia coli O157:H7 infection in humans. Ann Intern Med. 1995;123:698–714. doi: 10.7326/0003-4819-123-9-199511010-00009. [DOI] [PubMed] [Google Scholar]
- 40.Tetart F, Desplats C, Krisch H M. Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: recombination between conserved motifs swaps adhesin specificity. J Mol Biol. 1998;282:543–556. doi: 10.1006/jmbi.1998.2047. [DOI] [PubMed] [Google Scholar]
- 41.Tommassen J, van der Ley P, van Zeijl M, Agterberg M. Localization of functional domains in E. coli K-12 outer membrane porins. EMBO J. 1985;4:1583–1587. doi: 10.1002/j.1460-2075.1985.tb03820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanderslice R W, Yegian C D. The identification of late bacteriophage T4 proteins on sodium dodecyl sulfate polyacrylamide gels. Virology. 1974;60:265–275. doi: 10.1016/0042-6822(74)90384-5. [DOI] [PubMed] [Google Scholar]
- 43.Yu F, Mizushima S. Roles of lipopolysaccharide and outer membrane protein OmpC of Escherichia coli K-12 in the receptor function for bacteriophage T4. J Bacteriol. 1982;151:718–722. doi: 10.1128/jb.151.2.718-722.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu S, Ding H, Seah J, Wu K, Chang Y, Chang K S, Tam M F, Syu W. Characterization of a phage specific to hemorrhagic Escherichia coli O157:H7 and disclosure of variations in host outer membrane protein OmpC. J Biomed Sci. 1998;5:370–382. doi: 10.1007/BF02253447. [DOI] [PubMed] [Google Scholar]
- 45.Yu S L, Chou M J, Tam M F, Lee T H, Syu W J. Expression and antigenicity of human immunodeficiency virus type-1 transmembrane protein gp41 in insect cells. Biochem Biophys Res Commun. 1993;191:207–213. doi: 10.1006/bbrc.1993.1204. [DOI] [PubMed] [Google Scholar]
- 46.Zhao S, Meng J, Doyle M P, Meinersman R, Wang G, Zhao P. A low molecular weight outer-membrane protein of Escherichia coli O157:H7 associated with adherence to INT407 cells and chicken caeca. J Med Microbiol. 1996;45:90–96. doi: 10.1099/00222615-45-2-90. [DOI] [PubMed] [Google Scholar]