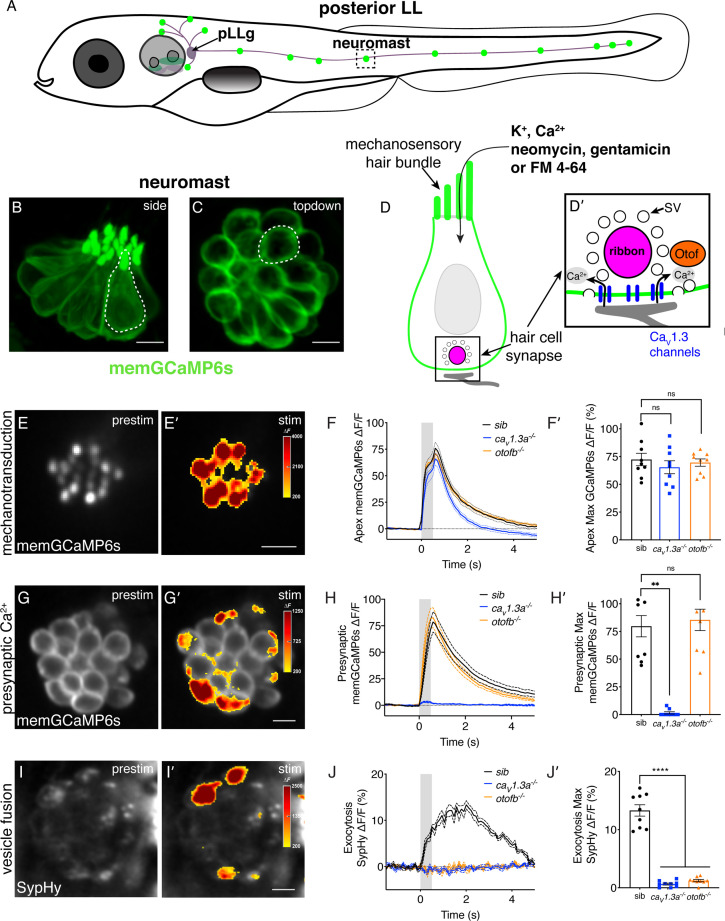
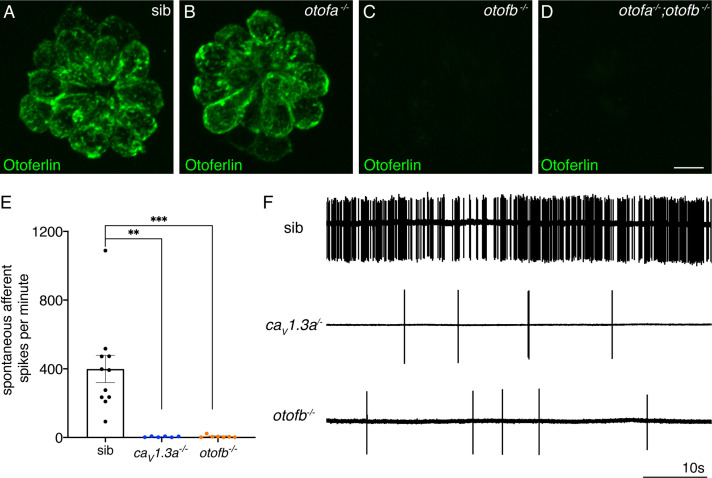
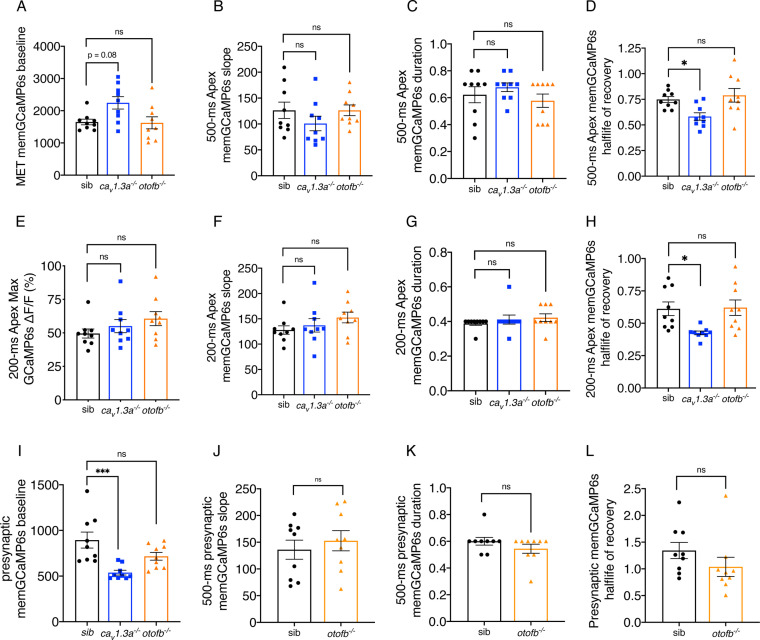
Figure 1. Genetic basis of neurotransmission in the zebrafish posterior-lateral line.
(A) Cartoon drawing of 5 dpf zebrafish larva with the posterior lateral line (LL) highlighted. The LL is made up of clusters of hair cell clusters called neuromasts (green dots). Neuromasts are innervated by neurons that project from the posterior LL ganglion (pLLg). (B–C) Side view (B) and top-down view (C) of neuromast from Tg[myo6b:memGCaMP6s]idc1 fish where the hair-cell membrane is labeled. White dotted line demarcates a single hair cell in each image. (D-D’) Cartoon schematic of a side view of a hair cell. At the apex is the mechanosensory hair bundle. The primary pathway of entry of neomycin, gentamicin, and FM 4–64 is through mechanotransduction channels in the mechanosensory hair bundle. At the base of the hair cell is the ribbon synapse. The presynapse or ribbon (magenta) is surrounded by synaptic vesicles (SV, white circles). When mechanotransduction channels are activated, an influx of cations including calcium enters the hair bundle. Hair bundle activation leads to opening of Cav1.3 voltage-gated calcium channels (blue) and presynaptic calcium influx. The calcium sensor Otoferlin (orange) facilitates fusion by coupling calcium influx with the exocytosis of SVs and the release of glutamate onto the innervating postsynaptic afferent terminal (gray). (E-E’) The spatial patterns of the evoked calcium influx (GCaMP6s ΔF, indicated via the heatmaps) into sibling hair bundles (E’) compared to prestimulus (C). (F-F’) Average traces (F) and dot plots show that the average magnitude of apical (F’) ΔF/F GCaMP6s signals in mechanosensory hair bundles is not different in cav1.3a-/- and otofb-/- mutants compared to siblings. (G-G’) The spatial patterns of the evoked calcium influx (GCaMP6s ΔF, indicated via the heatmaps) at wildtype presynapses (G’) compared to prestimulus (G). (H-H’) Average traces (H) and dot plots show that the average magnitude of presynaptic (H’) ΔF/F GCaMP6s signals is absent in cav1.3a-/- but unaltered in otofb-/- mutants compared to siblings. (I-I’) The spatial patterns of evoked exocytosis (SypHy ΔF, indicated via the heatmaps) at sibling presynapses (I’) compared to prestimulus (I). (J-J’) Averaged traces (J) and dot plots show that presynaptic (J’) ΔF/F SypHy signals are absent in cav1.3a-/- and in otofb-/- mutants. The fluid-jet stimulus depicted as a gray box in F, H, and J. Each point in the dot plots represents one neuromast. All measurements were performed in mature neuromasts at 5–6 dpf on 3 animals and 9 neuromasts per genotype. Error bars: SEM. A one-way AVOVA with a Dunnett’s correction for multiple tests was used in F’ and J’, and a Kruskal-Wallis test with a Dunn’s correction for multiple tests was used in H’. ** p<0.01, **** p<0.0001. Scale bar = 5 µm.