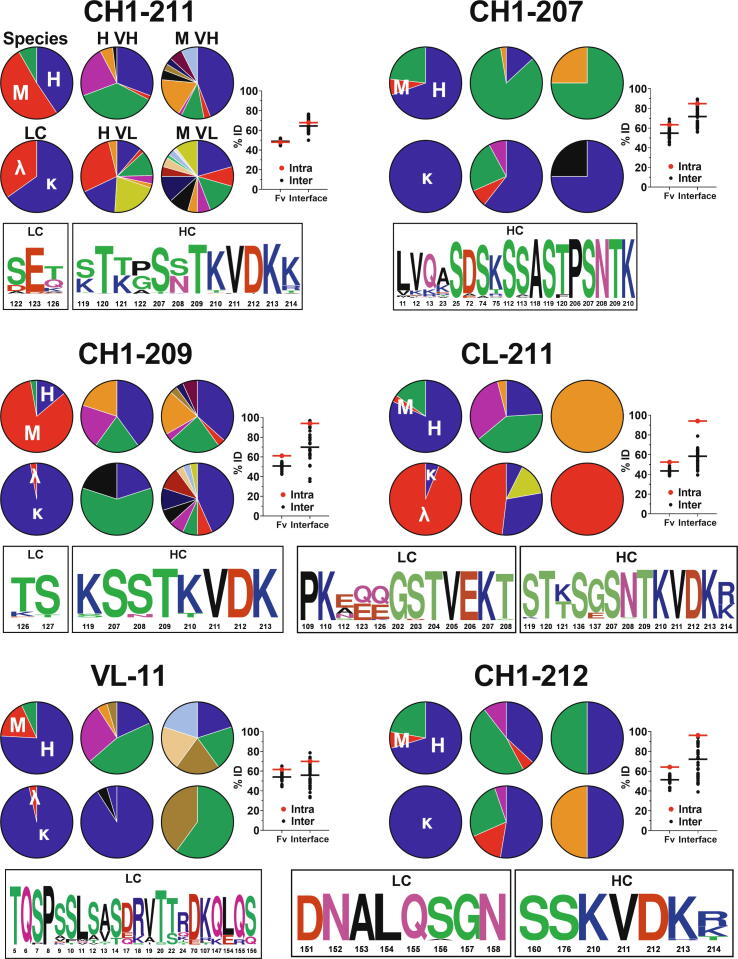
Fig. 4.
Interface profiles of the six most prevalent clusters. For all interfaces, the upper left-most pie chart provides the sequence distribution of cluster members based on species: human (blue, H), mouse (red, M), and other (e.g., rat, rabbit, etc., green). The lower left-most pie chart provides the sequence distribution of cluster members based on light chain type: kappa (blue, κ), and lambda (red, λ). The four right-most pie charts provide the sequence distribution of human VH (H VH), human VL (H VL), mouse VH (M VH), and mouse VL (M VL) subgroups. The labels above the pie charts in the upper left profile (CH1-211) are not repeated in the other charts for visual simplicity. The %ID plot on the right provides the intra (red) versus inter (black) cluster sequence identity for both the entire variable region (Fv) as well as only those residues at the interface as shown in the sequence logo and Supplementary Table S2. For the Fv, these values reflect the mean pairwise identities for all VH and VL sequences within the cluster (intra) versus the mean pairwise identities between each member of the cluster and all other members of all other clusters (inter). A similar comparison is made for each interface, where %ID reflects the mean pairwise identity of the residues at the interface of a given cluster aligned with those same residues for each member of the cluster (intra) versus all other members of all other clusters (inter). The sequence logo at the bottom provides weighted sequence composition at interface residues for members of the designated cluster, with numbering according to Kabat and EU conventions for the Fv and constant regions, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)