Fig. 5. Loss of interaction with c-Jun affects a subset of PU.1 target genes characterized by PU.1 sites lacking adjacent AP-1 site.
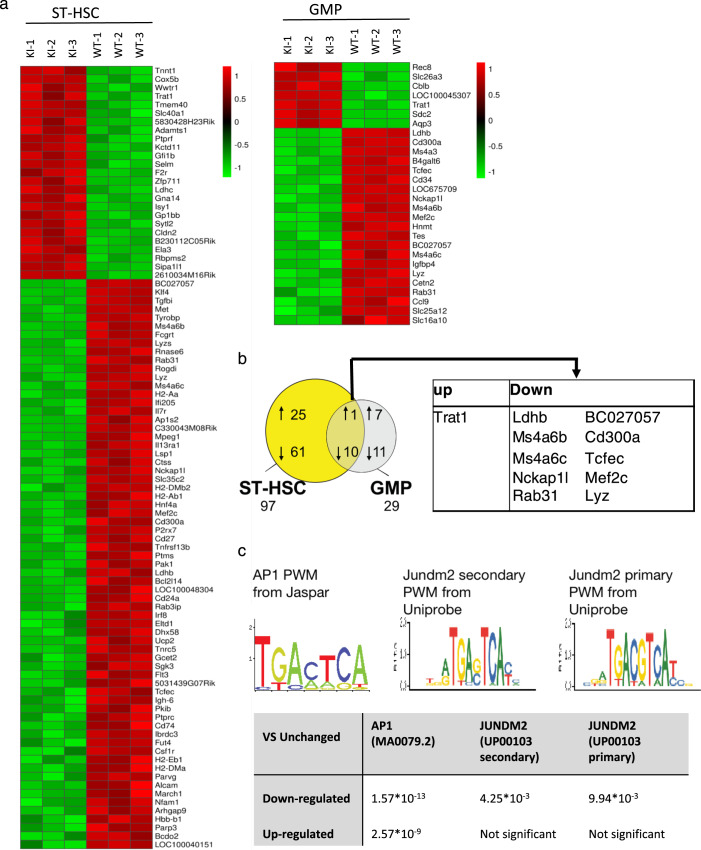
a Genome-wide mRNA expression profiles of ST-HSC (CD150− CD48− LSK, left) and GMP (lin− ckit+Sca1− CD16/32hi CD34+, right) were generated using Affymetrix MG430v2 microarrays. Hierarchical clustering of genes differentially expressed in PU.1 KI cells. Only genes with a log2 fold change >2.0 were considered differentially expressed. b Venn diagram showing the overlap of differentially expressed features between wild-type and mutant samples. Numbers below the populations correspond to the sum of the up- and downregulated genes in the mutant population compared to the wild-type counterpart. The genes differentially expressed in both populations are listed in the table below the diagram. c AP-1 consensus motifs found in unaffected gene promoters. Sequence logos that correspond to enriched sequence elements, as identified by de novo motif analysis in PU.1 ChIP-seq regions in genes with unchanged expression in the PU.1 mutant samples are shown. The table lists the statistical significance of the occurrence of the motifs. Consensus AP-1 motifs as well as a closely related motif with an additional base at the core of the motif corresponding to the Jundm2 consensus are shown.