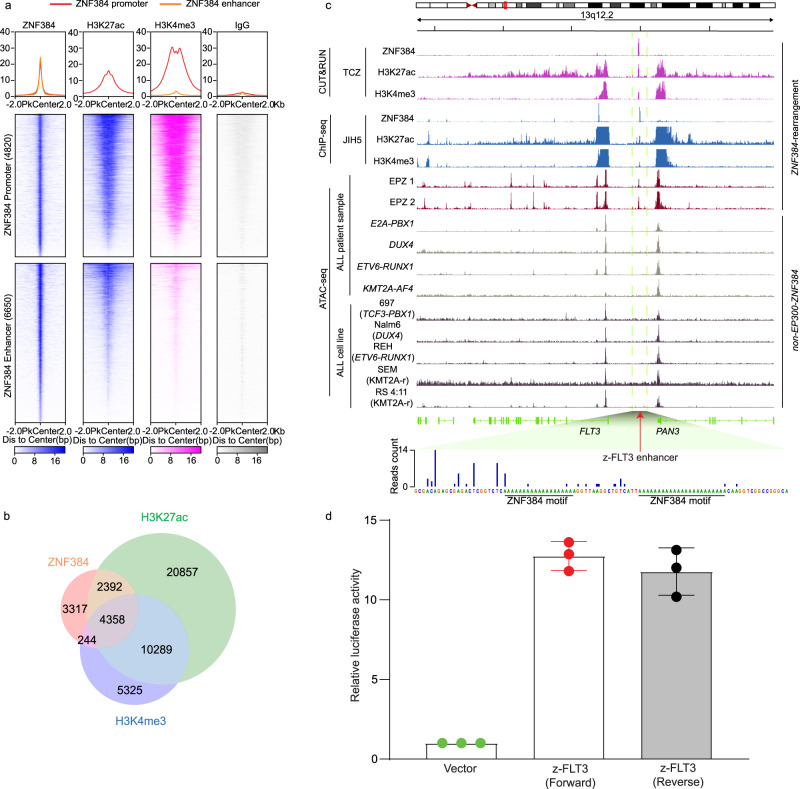
Fig. 2. ZNF384 fusion-specific activation of a distal enhancer at the FLT3 locus in ALL.
a, b ZNF384 CUT&RUN (Cleavage Under Targets & Release Using Nuclease) assay using a TCF3-ZNF384 ALL PDX sample (ID TCZ). In total, 10,470 peaks were identified, with 4820 in promoter regions (±2 kb from TSS) and the other 6650 in enhancers. 6750 peaks overlapped with H3K27ac peaks, while 4602 peaks overlapped with H3K4me3. c CUT&RUN and ATAC-seq (Assay for Transposase-Accessible Chromatin using sequencing) of ALL cell lines and primary ALL blast samples with or without ZNF384-rearrangements. Each track represents a type of assay in a given sample as indicated on the left. As shown in the CUT&RUN tracks, a unique ZNF384 binding site was observed 25 kb upstream of FLT3 with prominent H3K27ac and H3K4me3 marks. This peak also overlapped with an open chromatin region identified by ATAC-seq in EP300-ZNF384 ALL primary samples (n = 2). In contrast, this region was void of ATAC-seq signals in ALL cell lines or ALL primary samples of other subtypes. Bottom panel: two ZNF384 binding motifs (AAAAAAAA) were identified by footprint analysis using the ZNF384 CUT&RUN data derived from TCF3-ZNF384 ALL PDX cells. d Enhancer activity of z-FLT3 enhancer was confirmed by luciferase assay. A 500 bp segment (hg38, chr13:28,124,365–28,124,868) covered by this z-FLT3 enhancer increased transcription activity by 12.7-fold compared to vector control in JIH5 cells. Enhancer activity was normalized to empty vector control. Data are shown as mean values ± SEM of three biological replicates (center of the error bar) and the results are representative of three independent experiments. Source data are provided as a Source Data file.