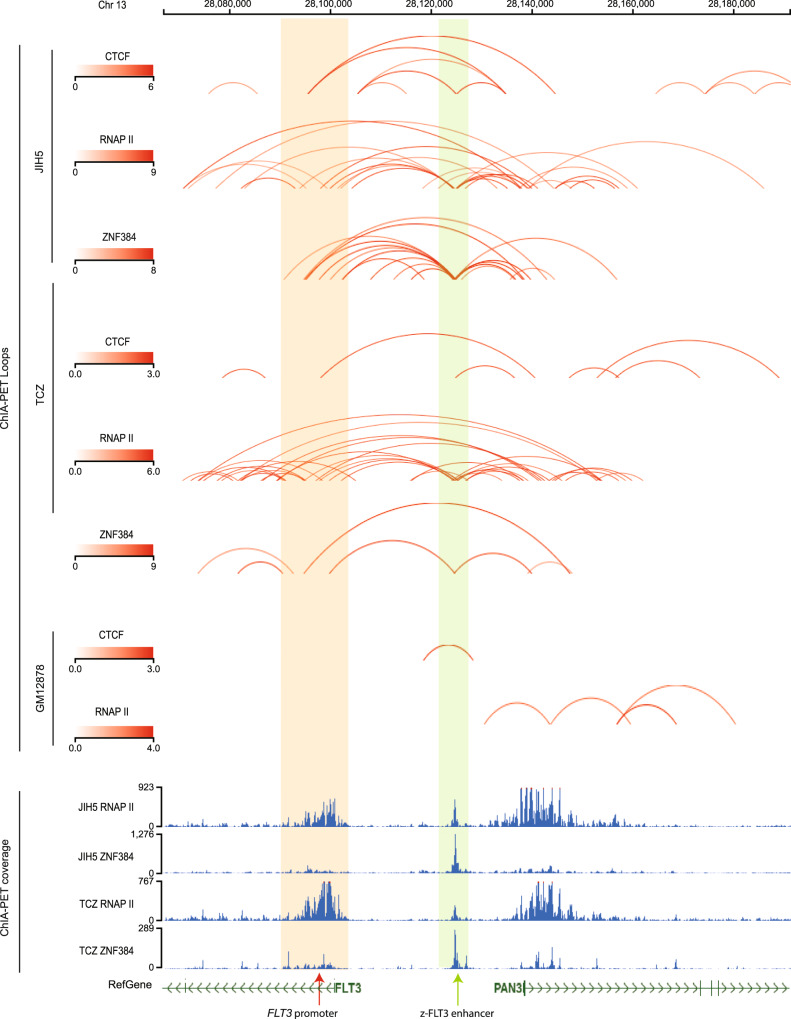
Fig. 3. 3D chromatin interaction at 5′-UTR of FLT3 in ZNF384-r ALL.
ChIA-PET (chromatin interaction analysis with paired-end tags) (CTCF and RNAP II) was performed in both JIH5 cells and an ALL PDX sample harboring the TCF3-ZNF384 fusion. As shown in the top four panels, DNA looping mediated by CTCF and RNAP II was detected between ZNF384-binding site (“z-FLT3 enhancer”) and FLT3 promoter, indicated by shaded box. In addition, we also performed ZNF384 ChIA-PET in both JIH5 cells and TCF3-ZNF384 ALL PDX sample to map ZNF384-mediated chromatin interactions, in comparison to lymphoblastoid cell line GM12878 which does not harbor ZNF384 fusion gene. Red curves indicated chromatin interactions called using ChIA-PIPE (paired-end tag reads ≥2) and were highlighted in red in Supplementary Data 1 and 2. Additionally, the bottom panels show protein binding sites (ZNF384 and RNAP II) identified from inter-ligation and self-ligation reads of ChIA-PET using MACS2 calling algorithm. Overlap of ZNF384 binding sites and chromatin looping anchors suggests enhancer-promoter interaction specifically mediated by ZNF384 (indicated by green and red arrows).