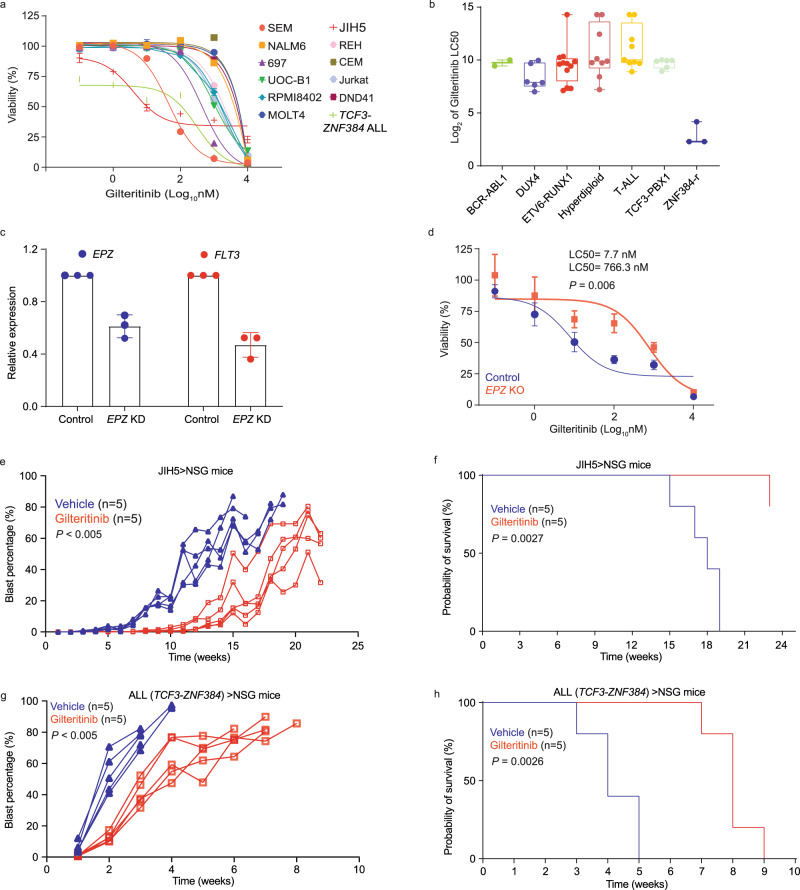
Fig. 4. Anti-leukemia effect of gilteritinib in vitro and in vivo.
a In vitro sensitivity of a panel of ALL cell lines as well as PDX-derived ALL cell with TCF3-ZNF384 fusion to gilteritinib was determined using MTT assay. Data are shown as mean % viability relative to vehicle ± SEM of three biological replicates (center of the error bar) and results are representative of three independent experiments. b Ex vivo sensitivity of a panel of 47 primary ALL cases including three with EP300-ZNF384 fusion as well as those with ETV6-RUNX1, DUX4-r, BCR-ABL1, hyperdiploidy, and T-ALL. Box plots show summary of data in terms of minimum, maximum, median, and first and third quartiles. Each data point represents two technical replicate. c, d Expression of EPZ (EP300-ZNF384) fusion gene was downregulated by CRISPR Cas9 editing in JIH5 cells, which led to decreased expression of FLT3 and drug sensitivity to gilteritinib. Data are shown as mean values ± SEM of three biological replicates (center of the error bar) and results are representative of three independent experiments. P values (P = 0.006) were estimated using two-sided t test. e–h Gilteritinib efficacy was evaluated in vivo using xenograft models. e, f Show leukemia progression, and survival in mice transplanted with JIH5 cells. g, h Describe results of mice transplanted with PDX-derived ALL cells with TCF3-ZNF384 fusion. Gilteritinib was given daily at a dosage of 10 mg/kg, and therapy started three days following leukemia engraftment. Leukemia burden was monitored weekly, and P values (P < 0.005) were estimated using a two-sided analysis of deviance based on mixed effect models with cubic splines. Leukemia-free survival was plotted as Kaplan–Meier curves and P values were estimated using two-sided log-rank test (P = 0.0027 for f, P = 0.0026 for h). Source data are provided as a Source Data file.