Abstract
Circular RNA (circRNA) is characterized by a specific covalently closed ring structure. The back-splicing of precursor mRNA is the main way of circRNA generation, and various cis/trans-acting elements are involved in regulating the process. circRNAs exhibit multiple biological functions, including serving as sponges of microRNAs, interacting with proteins to regulate their stabilities and abilities, and acting as templates for protein translation. Autophagy participates in many physiological and pathological processes, especially it plays a vital role in tumorigenesis and carcinoma progression. Increasing numbers of evidences have revealed that circRNAs are implicated in regulating autophagy during tumor development. Until now, the roles of autophagy-associated circRNAs in carcinoma progression and their molecular mechanisms remain unclear. Here, the emerging regulatory roles and mechanisms of circRNAs in autophagy were summarized. Furtherly, the effects of autophagy-associated circRNAs on cancer development were described. We also prospected the potential of autophagy-associated circRNAs as novel therapeutic targets of tumors and as biomarkers for cancer diagnosis and prognosis.
Subject terms: Tumour biomarkers, Oncogenes
Facts
circRNAs are implicated in diverse physiological processes, including autophagy. They are also associated with the pathogenesis of numerous diseases, such as cardiovascular diseases, neurological disorders, and cancer.
circRNAs have both stimulatory and inhibitory effects on autophagy.
circRNA-mediated autophagy plays vital roles in multiple aspects of tumor progression, especially the development of chemoresistance in cancer.
The expression patterns of circRNAs are associated with the clinicopathological characteristics of cancer.
circRNAs are considered to represent novel therapeutic targets and potentially useful diagnostic and prognostic biomarkers of cancer.
Open questions
How do circRNAs regulate autophagy in the context of cancer?
How does circRNA-mediated autophagy affect tumorigenesis and cancer development?
Should we promote or inhibit circRNA-mediated autophagy to suppress cancer progression?
Whether targeting autophagy-associated circRNAs can be novel strategies for cancer treatment?
Introduction
Circular RNA (circRNA) is a class of endogenous RNA molecules with special covalent closed-loop structure. CircRNAs are mainly created from precursor mRNA (pre-mRNA) back-splicing, and they have been identified in a variety of eukaryotes and viruses [1, 2], and exhibit developmental-stage-specific and tissue-specific expression patterns [3, 4]. When they were first discovered nearly 50 years ago, these RNA molecules received little attention and were thought to be derived from errors in the splicing of mRNA and thus lacking in biological importance. However, recently researchers have found that circRNAs are implicated in diverse physiological processes, including autophagy [5] and immunity [6, 7]. They are also associated with the pathogenesis of numerous diseases, such as cardiovascular diseases [8], neurological disorders [9], chronic inflammatory diseases [10, 11], and cancer [12, 13]. The roles played by circRNAs in tumorigenesis and cancer progression have also recently been revealed. Numerous dysregulated circRNAs have been identified in diverse carcinomas, such as colorectal cancer (CRC) [14, 15], gastric cancer (GC) [16–18], basal cell carcinoma [19, 20], and hepatocellular carcinoma (HCC) [21, 22], and their expression was related to the clinicopathological features of cancer patients. We previously analyzed the expression profiles of circRNAs in five pairing GC and corresponding adjacent tissues, using circRNA sequencing with linear RNA depletion [16]. We totally identified 45,783 circRNAs from these samples, including 79% exonic, 1% intronic, 4% intergenic, 15% sense overlapping, and 1% antisense[16]. These circRNAs may participate in GC development. It has become apparent that circRNAs play vital roles in cell death [23], chemoresistance [24], metastasis [25], the maintenance of cancer-initiating cells [26], immune evasion [27], and angiogenesis [28] in cancer. circRNAs usually show higher stability compared with that of their linear host gene, presumably due to their cyclic structures, which render them resistant to exonuclease-mediated degradation. Furthermore, some circRNAs are detectable in body fluids, including saliva, urine and peripheral blood, which suggests the possibility of their being used as non-invasive biomarkers [29]. Thus, circRNAs show great potential as diagnostic and prognostic biomarkers and as therapeutic targets for cancer therapy.
Autophagy, as a highly conserved catabolic process, plays key roles in maintaining a balance between cellular survival and death, which regulates the degradation and recycling of intracellular materials, including damaged organelles and misfolded proteins. It renders cells resistant to survival stress (such as nutritional deficiency and hypoxia), via providing materials and energy for synthesis of new cellular components and thereby restoring cellular homeostasis [30]. Three types of autophagy (macroautophagy, microautophagy, and chaperone-mediated autophagy) have been identified [31]. This review is focused on macroautophagy, characterized by the formation of autophagosomes that possess a double membrane structure. In macroautophagy (hereinafter referred to as autophagy), after being enveloped by autophagosomes, damaged cellular components are transported to lysosomes for degradation and recycling. To date, the role autophagy plays in cancer development remains controversial. On one hand, autophagy suppresses tumorigenesis by maintaining genome stability and homeostasis of cellular metabolism. On the other hand, it participates in reprogramming cellular microenvironment following the establishment of cancer and protects cancer cells from diverse survival stresses [32]. Autophagy also helps cancer cells escape from anti-tumor immune responses mediated by natural killer cells and cytotoxic T-lymphocytes[33]. Furthermore, increasing evidence has revealed the double role circRNAs play in autophagy regulation. Some circRNAs have been found to promote autophagy. A study by Yang and colleagues determined that circRHOBTB3 functions as a microRNA (miRNA) sponge, and facilitates autophagy via circRHOBTB3/miR-600/NACC1 axis, leading to increased cellular proliferation in pancreatic ductal adenocarcinoma (PDAC) [34]. Conversely, other circRNAs negatively regulate autophagy. circUBE2Q2 was shown to suppress STAT3-mediated autophagy via sponging miR-370-3p but promote tumorigenicity in GC [35]. Thus, to develop novel circRNA-based therapeutic strategies for cancer, the mechanisms of how circRNAs regulate autophagy and how circRNA-regulated autophagy affects tumorigenesis and progression must be urgently clarified.
Here, the emerging findings in relation to the regulatory roles circRNAs play in autophagy are summarized and these molecules’ effects on cancer development and progression are described. Furtherly, the potential of circRNAs as novel biomarkers of cancer diagnosis and prognosis and therapeutic targets for tumor treatment is also prospected.
circular RNA
circular RNA biogenesis
The biogenesis of circRNAs is complicated, but they are primarily produced from pre-mRNA back-splicing, which is characterized by direct binding of a 5’ downstream splice site (donor) to a 3’upstream splice stie (acceptor) via a phosphodiester bond (3’–5’)[36]. Based on the origin of circRNAs, they are mainly classified into 3 groups: exon-derived circRNA (EcRNA), intron-derived circRNA, including intronic circRNAs derived from pre-mRNA (ciRNA) and those derived from pre-tRNA (tricRNA), and exon–intron circRNA (EIciRNA) [12]. Various cis-/trans-acting factors is implicated in the regulation of circRNA generation (Fig. 1). Some intronic complementary sequences, including short inverted repeats (such as Alu repeats) [37] and non-repetitive sequences, can act as cis-acting elements to facilitate circRNA biogenesis. The circularization of exons can be mediated by these complementary sequences located in flanking introns, by forming an intramolecular hairpin structure to close the distance between the 5′ and 3′ splice sites. Short inverted repeats (approximately 30–40 nt) are sufficient for circularization; however, the sequences of the repeats affect circularization efficiency. Some sequences of low complexity, such as poly(A) tracts, suppress circularization [37]. Moreover, as trans-acting factors, some RNA-binding proteins (RBPs), including muscleblind [36] and Quaking (QKI) [38], facilitate circRNA production by binding to specific intronic motifs. QKI binds to theses intronic elements flanking circRNA-forming exons, then brings the splice sites into proximity via self-dimerization. Additionally, Nuclear factor 90 (NF90) and NF110, the immune factors mediating host immune responses to viral infections, also regulate the circularization of circRNA [39]. They contain double stranded (ds) RNA-binding domains and facilitate circRNA generation via binding to RNA pairs in flanking introns and stabilizing the base-pairing. Conversely, some RPBs, such as DExH-box helicase 9, suppress circRNA biogenesis via destabilizing the base-pairing of intronic elements.
Fig. 1. circRNA biogenesis.
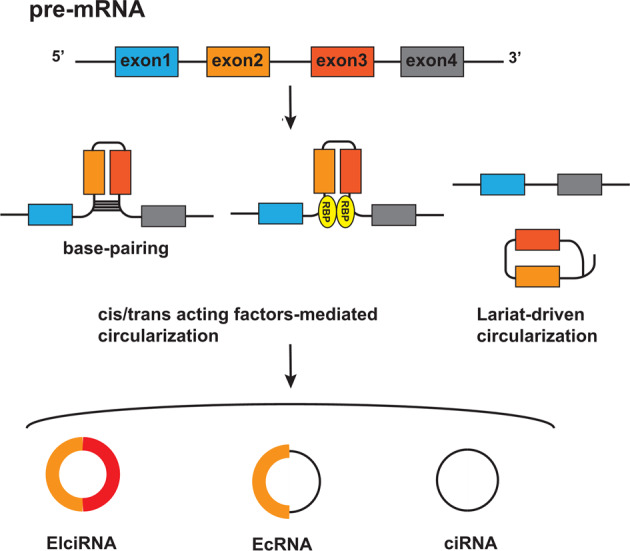
CircRNAs are mainly generated from the back-splicing of pre-mRNA. Cis/trans-acting factor-mediated circularization (left). As cis-acting elements, some intronic complementary sequences facilitate circularization via the formation of an intramolecular hairpin structure, by base-pairing to close the distance between the 5′ and 3′ splice sites. Meanwhile, as trans-acting factors, some RNA-binding proteins (RBPs) facilitate the production of circRNAs by binding to specific intronic motifs to bring the splice sites into proximity via self-dimerization. Lariat-driven circularization (right). In exon-skipping events, the skipped exons and introns are spliced out from the pre-mRNA and form a lariat structure, which is further spliced to form circRNAs. circRNAs can be mainly divided into three groups, based on their origin: exon-derived circRNA (EcRNA), exon–intron circRNA (EIciRNA), and intron-derived circRNA from pre-mRNA (ciRNA).
Another model for circRNA biogenesis is called “Lariat-driven circularization” [12]. During exon-skipping events, a lariat structure is formed via splicing the skipped exons and introns out from the pre-mRNA, which is further spliced to remove intronic sequences, resulting in the generation of an EcRNA. Alternatively, a lariat structure may be formed during intron removal from pre-mRNAs, which is further spliced for ciRNA generation.
The main mechanism underlying circRNA functions
circRNAs exhibit multiple functions in physiological and pathological processes. The sequence, secondary structure, post-transcriptional modifications and cellular location of circRNAs are related to their functions.
microRNA (miRNA) sponge
The most well-studied function of circRNAs is known as miRNA sponging. Some circRNAs negatively regulate miRNA-mediated gene silencing by acting as competitive endogenous RNAs (ceRNAs) (Fig. 2A). circHERC4 was recently found to exhibit oncogenic effects in CRC, by binding to and inactivating the tumor suppressor, miR-556-5p [40]. circDOCK1 also acts as a miRNA sponge and promotes tumorigenesis in osteogenic sarcoma, via the circDOCK1/ miR-339-3p/IGF1R axis [41]. Those circRNAs with function as miRNA sponges harbor miRNA binding sites (6-, 7-, 8-mer), complementary to miRNA seed sequences. However, the miRNA binding sites in most circRNAs are very few, and the expression level of circRNAs is relatively low compared with their corresponding miRNAs. These findings mean that the ceRNA hypothesis remains controversial [42].
Fig. 2. The main mechanisms underlying the functions of circRNA.
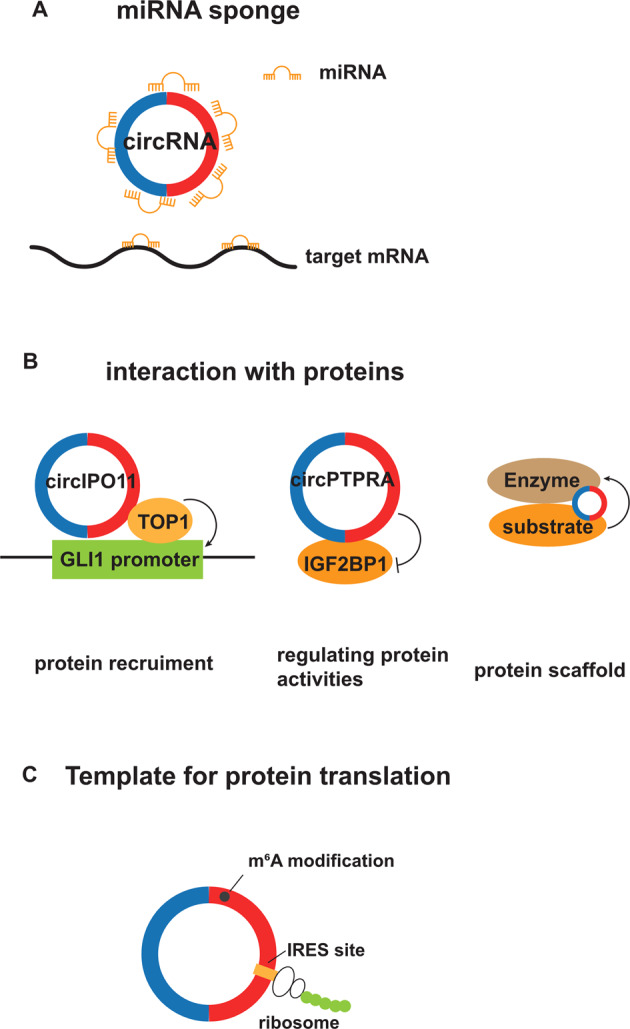
A miRNA sponge. Some circRNAs negatively regulate miRNA-mediated gene silencing by competitively binding to miRNA and releasing its target mRNA. B Interaction with proteins. CircIPO11 interacts with topoisomerase 1 (TOP1) and then recruits it to the GLI1 promoter to trigger protein expression. circPTPRA interacts with IGF2BP1 and inhibits its function as an m6A reader. As protein scaffolds, some circRNAs interact with an enzyme and its substrate and then facilitate the association between them. C As a template for protein translation. Some circRNAs containing internal ribosome entry sites (IRES) and those with N6-methyladenosine (m6A) modification can serve as templates for protein translation.
Interactions with proteins
Some circRNAs bind to proteins and recruit them to certain subcellular compartments (Fig. 2B) [26, 43, 44]. Gu and colleagues described a circRNA, circIPO11, that is required to maintain the self-renewal of cancer-initiating cells in HCC [26]. circIPO11 was shown to trigger the expression of GLI family zinc finger protein1 via recruiting topoisomerase 1 to its promoter, and result in the activation of Hedgehog signaling. circMYH9, an intron-derived circRNA, was found to recruit hnRNPA2B1 in the nucleus and bound to p53 pre-mRNA to maintain its stability. In this way, circMYH9 promotes CRC cell proliferation in a p53-dependent manner [44]. Some circRNAs interact with proteins and suppress their activities [45–48]. circPTPRA was shown to suppress the progression of bladder cancer, through its interaction with IGF2BP1, an N6-methyladenosine (m6A) reader, and blocking the IGF2BP1-mediated recognition of m6A-modified RNAs [47]. Furthermore, some circRNAs function as protein scaffolds to affect interactions among proteins [49, 50]. As an example, circRNA-DOPEY2 facilitates the association between the E3 ligase TRIM25 and its substrate cytoplasmic polyadenylation element binding protein (CPEB4), leading to increased CPEB4 degradation [50]. As a result, the circRNA promotes chemosensitivity in esophageal cancer cells.
Translation
Although circRNAs were previously identified as non-coding RNAs, because they lacked a 5′ cap structure and a 3′ poly-A tail, recently some circRNAs containing internal ribosome entry sites (IRES) have been revealed to serves as templates for protein synthesis (Fig. 2C). We recently described AXIN1–295aa as a novel oncogenic factor in GC, which is encoded by circAXIN1with an IRES element [16]. We determined that this protein promotes GC development by interacting with APC and promoting the activation of Wnt/β-catenin signaling. Several circRNAs with m6A modification also exhibit protein-coding potential. Li et al. identified a novel oncogenic circRNA, circARHGAP35, which harbors an open reading frame (ORF) with an m6A-modified start codon and encodes a protein, referred to as circARHGAP35 protein. They further determined that the circRNA-derived protein shows antithetical expression and function compared with its linear partner and enhances the development of HCC and CRC via the interaction with TFII-I protein [51]. Furthermore, circDIDO1 with IRES, ORF, and m6A modification encodes a novel tumor suppressor protein, which suppresses GC progression by interacting with peroxiredoxin 2 (PRDX2) and poly ADP-ribose polymerase 1 (PARP1), affecting the activity or stability of these proteins [52].
Autophagy
circRNAs have both stimulatory and inhibitory effects on autophagy, and they mainly regulate the process of autophagy via ceRNA mechanisms. circRNA-mediated autophagy is implicated in cancer development and plays vital roles in cell proliferation, metastasis, chemoresistance, and apoptosis.
The roles played by circRNAs in autophagic processes
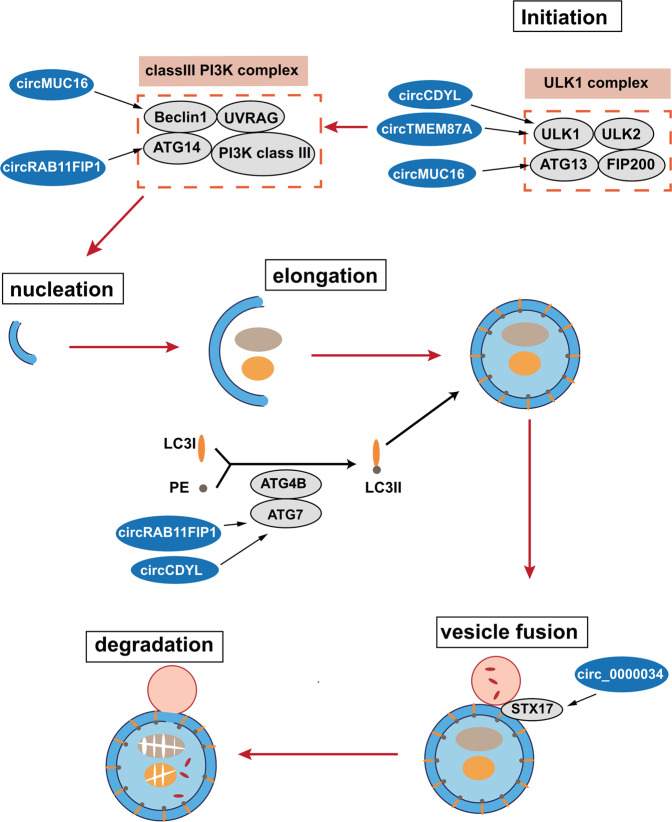
There are mainly five stages in the autophagic processes: initiation, autophagosome nucleation, autophagosome membrane elongation, the fusion between the autophagosome and a lysosome, and the degradation of the autophagic cargo [31] (Fig. 3). circRNAs are implicated in these processes via interacting with autophagy-related proteins or regulating their expression. During the initiation stage, a unc-51-like kinase 1 (ULK1) complex, composed of ULK1, ULK2, autophagy-related 13 (ATG13), and FIP200 is activated [31]. Several circRNAs are associated with the initiation stage. circCDYL and circTMEM87A regulate the expression of ULK1 via acting as the sponges of miR-1275 and miR-142-5p, respectively, and further promote autophagy [53, 54]. circMUC16 promotes ATG13 expression and facilitates autophagy via directly binding to ATG13 in epithelial ovarian cancer (EOC) [55]. Then, the ULK1 complex induces the activation of a class III PI3K complex (containing Beclin1, ATG14, UVRAG, and class III PI3K), which mediates autophagosome nucleation. During this process, circMUC16 enhances autophagy via the circMUC16/ miR-199a-5p/Beclin1 axis [55]. circRAB11FIP1 regulates ATG14 expression by sponging miR-129 [56]. The complex of ATG5-ATG12 mediates the elongation of the autophagosome membrane, via conjugating with ATG16 protein. Meanwhile, LC3-II is formed via the conjugation of LC3-I with lipid phosphatidylethanolamine (PE), followed by being recruited to the membrane of autophagosome. This process is mediated by the ATG4B-ATG7 complex. circRAB11FIP1 and circCDYL are reported to regulate ATG7 expression via serving as the sponges of miR-129 and miR-1275, respectively, and promote autophagy [53, 56]. Subsequently, the fusion between autophagosomes and lysosomes is facilitated by the SNARE protein syntaxin 17 (STX17), resulting in the degradation of autophagic cargoes. Circ_0000034 is implicated in the process by regulating the miR-361-3p/STX17 axis [57].
Fig. 3. The role of circRNAs in the process of autophagy.
The autophagic processes can be divided into five stages: initiation, autophagosome nucleation, the elongation of the autophagosome membrane, the fusion of the autophagosome with a lysosome, and the degradation of the autophagic cargo. circRNAs are involved in these processes by interacting with autophagy-related proteins or regulating their expression.
The multiple signaling pathways associated with circRNA-mediated regulation of autophagy
circRNAs regulate autophagy via a diverse range of signaling pathways, as described below (Fig. 4).
Fig. 4. Multiple signaling pathways are implicated in the regulation of autophagy by circRNAs.
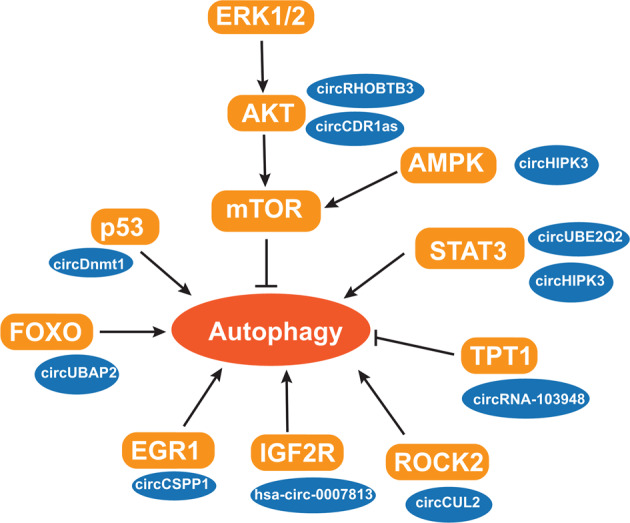
circRNAs participate in multiple signaling pathways to regulate autophagy, by acting as miRNA sponges or through interactions with proteins.
Signal transducer and activator of transcription 3 (STAT3)
STAT3 signaling, a stress response pathway, has been reported to participate in regulating autophagy [58]. STAT3 proteins located in various subcellular compartments can affect the autophagic process in diverse ways [58]. Phosphorylated STAT3 proteins form dimers and enter the nucleus to regulate the expression of autophagy-associated genes, including Bcl-2, BECN1, and PIK3C3. Cytoplasmic unphosphorylated STAT3 suppresses autophagy by sequestering FOXO1, FOXO3, and EIF2AK2. circUBE2Q2 was shown to suppress miR-370-3p/STAT3-mediated autophagy and promote GC development. Its knockdown was found to significantly decrease STAT3, p-STAT3, and Bcl-2 levels, resulting in increased autophagy [35]. Chen et al. also identified a circRNA, circHIPK3, as a key autophagy regulator in lung cancer [59]. circHIPK3 promotes cancer progression and suppresses autophagy, partially by sponging miR-124-3p and regulating the expression of its target, STAT3.
Mammalian target of rapamycin (mTOR)
mTOR is a key regulator of autophagy. As a serine–threonine kinase, mTOR forms an mTORC1 complex with mLST8 and RAPTOR proteins, and inhibits the ULK1 complex by inducing the phosphorylation of the complex components, including ULK1/2 and ATG13 [60]. Several signaling pathways participate in autophagy via the regulation of mTOR activity, including the adenosine monophosphate-activated protein kinase (AMPK), phosphoinositide 3-kinase (PI3K)/protein kinase (AKT) and extracellular-signal-regulated kinase (ERK) signaling pathways [61]. Yang et al. identified a pro-autophagic circRNA, circRHOBTB3, in PDAC [34]. This circRNA functions as a miRNA sponge, and relieves miR-600-mediated inhibition of expression of NACC1, and then facilitates autophagy via suppresses the Akt/mTOR signaling. circCDR1as is reported to promote autophagy and the development of oral squamous cell carcinoma (OSCC), by sponging miR-671-5p [62]. It enhances hypoxia-induced autophagy via the upregulation of the AKT and ERK1/2 pathways and inhibiting mTOR activity. Furthermore, circPAN3 was determined to facilitate autophagy and chemoresistance in acute myeloid leukemia through AMPK-mTOR signaling [63].
p53
The p53 pathway exhibits both promoting and inhibiting effects on autophagy, according to its cellular localization [64–66]. Nuclear p53 promotes autophagy via transactivating the expression of pro-autophagic proteins, such as Sestrin1 and Sestrin2 [67], AMPK [68, 69] and damage-regulated autophagy modulator (DRAM) [70, 71], while cytoplasmic p53 is involved in autophagy inhibition. circDnmt1 induces autophagy and increases the survival capacity of cells in breast cancer (BCa), by promoting the transport of p53 and AUF1 into cellular nucleus [15]. Nuclear p53 stimulates autophagy, while nuclear AUF1 increases the expression of Dnmt1, which inhibits p53 transcription.
Forkhead box class O (FOXO)
The effects of FOXO transcription factors on autophagy are also dependent on their cellular location. The cytoplasmic FOXOs interact with autophagy-associated proteins directly, while the nuclear ones regulate autophagy-related gene expression [72, 73]. Chen and colleagues demonstrated the pro-autophagic and oncogenic effects of circUBAP2 in CRC, which regulates the expression of miR-582-5p-targeted FOXO1 by sponging this circRNA [74]. circMRPS35 induces autophagy in osteosarcoma via interacting with KAT6B, and further promoting FOXO3 expression [75].
Other pathways associated with circRNA-mediated autophagy
Early growth response factor 1 (EGR1) was identified as a transcriptional regulator of autophagy, modulating the expression of autophagy-related genes [76]. CircCSPP1 facilitates autophagy and enhances the progression of prostate cancer (PCa), via the miR-520h/EGR1 axis [77]. Insulin-like growth factor 2 receptor (IGF2R) also regulates the autophagic processes. The loss of IGF2R results in lysosome dysfunction and autophagy inhibition [78]. hsa_circ_0007813 was shown to exhibit pro-autophagic and oncogenic effects on the development of bladder cancer, by modulating the expression of hsa-miR-361-3p-targeted IGF2R [79]. ROCK2 is also implicated in autophagy [80]. circCUL2 activates ROCK2-mediated autophagy via sponging miR-142-3p and then modulates chemoresistance in GC [81]. Furthermore, the tRNA splicing enzyme TPT1 has been reported to negatively regulate autophagy [82]. circRNA_103948 was found to inhibit autophagy in CRC by regulating the expression of miR-1236-3p-targeted TPT1 [83].
The diverse effects of circRNA-mediated autophagy in cancer
circRNA-mediated autophagy plays vital roles in multiple aspects of tumor progression, including cellular proliferation, invasion and metastasis. It is also associated with cancer escape mechanisms and renders cancer cells resistant to chemotherapy.
The role of circRNA-mediated autophagy in tumor development
circRNA-mediated autophagy can exhibit either promoting or inhibiting effects on cancer development across different tumor types (Table 1). In BCa, autophagy enhanced by circDnmt1 and circCDYL promotes the malignant progression of tumors [15, 53], while hsa_circ_0000515 knockdown induces autophagy and apoptosis but suppresses cellular proliferation and invasion in cervical cancer, suggesting an anti-tumor effect of autophagy [84]. circRHOBTB3-mediated autophagy was found to promote cell proliferation in PDAC [34]. It has even been noted that autophagy can have antithetical effects on cancer progression, even within the same tumor type. The silencing of circUBE2Q2 increased autophagy but suppressed glycolysis, cell proliferation, invasion, and migration in GC [35]. Meanwhile, circCUL2 was found to activate autophagy but inhibit tumorigenicity in GC [81]. These findings suggest that autophagy regulated by circUBE2Q2 and circCUL2 exhibits anti-tumor activity in GC, whereas circTMEM87A-mediated autophagy positively regulates GC progression [54]. Similarly, circUBAP2 promotes autophagic processes and enhances the development and metastasis of CRC [74], while circRNA_103948 suppresses autophagy and promotes the progression of CRC [83]. These findings indicate the opposing effects of autophagy on CRC progression. Therefore, the effects of autophagy on tumorigenesis and progression may be dependent on tumor types, the genetic context of cells, and specific types of cellular stresses. An understanding of the mechanisms underlying how circRNAs regulate autophagic processes and how autophagy affects tumorigenesis is thus important for the development of circRNA-based cancer therapy strategies.
Table 1.
The roles of autophagy-associated circRNAs in cancer development.
| circRNA | Roles in autophagy | Mechanism | Target | Cancer types | Roles in cancer development | Dysregulation in cancer(up/down) | References |
|---|---|---|---|---|---|---|---|
| circDnmt1 | Promote | Recruitment of proteins | P53 and AUF1 | Bca | Oncogene | Up | [15] |
| circCSPP1 | Promote | ceRNA | miR-520h/EGR1 | PCa | Oncogene | Up | [77] |
| circUBAP2 | Promote | ceRNA | miR-582-5p/FOXO1 | CRC | Oncogene | Up | [74] |
| circRHOBTB3 | Promote | ceRNA | miR-600/NACC1 | PDAC | Oncogene | Up | [34] |
| circRAB11FIP1 | Promote | ceRNA | miR-129/ATG7 and ATG14 | EOC | Oncogene | Up | [56] |
| circCDYL | Promote | ceRNA | miR-1275/ATG7 and ULK1 | BCa | Oncogene | Up | [53] |
| circMUC16 | Promote | ceRNA; Interaction with protein | miR-199a-5p/Beclin1 and RUNX1; ATG13 | EOC | Oncogene | Up | [55] |
| hsa_circ_0007813 | Promote | ceRNA | hsa-miR-361-3p/IGF2R | bladder cancer | Oncogene | Up | [79] |
| circTMEM87A | Promote | ceRNA | miR-142-5p/ULK1 | GC | Oncogene | Up | [54] |
| circCDR1as | Promote | ceRNA | miR-671-5p | OSCC | Oncogene | Up | [62] |
| circ_0000034 | Promote | ceRNA | miR-361-3p/STX17 | RB | Oncogene | Up | [57] |
| circMRPS35 | Promote | Interaction with protein | KAT6B/FOXO3 | osteosarcoma | Anti-oncogene | _ | [75] |
| circUBE2Q2 | Suppress | ceRNA | miR-370-3p/STAT3 | GC | Oncogene | Up | [35] |
| circHIPK3 | Suppress | ceRNA | miR-124-3p/STAT3 | NSCLC | Oncogene | _ | [59] |
| hsa_circ_0000515 | Suppress | ceRNA | miR-326/ELK1 | Cervical cancer | Oncogene | Up | [84] |
| circRNA_103948 | Suppress | ceRNA | miR-1236-3p/TPT1 | CRC | Oncogene | Up | [83] |
Bca breast cancer, PCa prostate cancer, GC gastric cancer, CRC colorectal cancer, PDAC pancreatic ductal adenocarcinoma, EOC epithelial ovarian cancer, NSCLC non-small cell lung cancer, OSCC oral squamous cell carcinoma, RB retinoblastoma.
The role of circRNA-mediated autophagy in chemoresistance of cancer cells
circRNAs are also implicated in the chemoresistance of tumor cells by regulating autophagy (Table 2). Cisplatin, a platinum-based reagent, has been widely used in therapy for solid cancers, such as ovarian, lung, colorectal, bladder, and head and neck cancers [85, 86]. However, chemoresistance often develops following cisplatin-based therapy, limiting its clinical utility [87]. circPARD3 facilitates chemoresistance of cancer cells to cisplatin in laryngeal squamous cell carcinoma via regulating PRKCI-AKT-mTOR signaling and then suppressing autophagy [88]. The silencing of circPARD3 increases the sensitivity of cancer cells to this drug. circCUL2 also affects the resistance of GC cells to cisplatin via modulating miR-142-3p/ROCK2-mediated autophagy [81]. Zhong and colleagues described a circRNA, circRNA_100565, associated with the resistance of non-small cell lung cancer (NSCLC) cells to cisplatin [89]. The upregulation of circRNA_100565 was observed in cisplatin-resistant NSCLC cell lines and tissues. Furtherly, this circRNA promotes autophagy and cell proliferation, but suppresses apoptosis via the miR-337-3p/ADAM metallopeptidase domain 28 (ADAM28) axis, resulting in the enhancement of cisplatin resistance. In addition, several circRNAs contribute to the resistance of cancer cells to apatinib, a selective inhibitor of vascular endothelial growth factor receptor 2 (VEGFR2), which also exhibits anti-tumor activity in solid tumors [90]. Ma et al. showed that circRACGAP1 is involved in the resistance of cancer cells to Apatinib in GC via regulating the miR-3657/ATG7-mediated autophagy. circRACGAP1 knockdown sensitized GC cells to the drug via suppressing autophagy [91]. Additionally, this circRNA regulates gefitinib resistance in NSCLC via targeting miR-144-5p/CDKL1. circRACGAP1 knockdown significantly increases the gefitinib sensitivity in NSCLC cells [92]. The upregulation of circ_0009910 was noted in the serum of imatinib-resistant patients with chronic myeloid leukemia (CML) [93]. circ_0009910 enhances the resistance of CML cells to imatinib via sponging miR-34a-5p and then regulating ULK1-mediated autophagy. Thus, the roles played by circRNAs in the chemoresistance of cancer cells suggest that targeting circRNA-mediated autophagy is a potential strategy to attenuate chemoresistance in patients with advanced cancer.
Table 2.
The roles of autophagy-associated circRNAs in chemoresistance.
| circRNA | Roles in autophagy | Mechanism | Target | Anti-cancer reagents | Cancer types | Roles in cancer development | Dysregulation in cancer(up/down) | References |
|---|---|---|---|---|---|---|---|---|
| circCUL2 | Promote | ceRNA | miR-142-3p/ROCK2 | Cisplatin | GC | Anti-oncogene | Down | [81] |
| circRNA_100565 | Promote | ceRNA | miR-337-3p/ADAM28 | Cisplatin | NSCLC | Oncogene | Up | [89] |
| circRACGAP1 | Promote | ceRNA | miR-3657/ATG7 | Apatinib | GC | Oncogene | Up | [91] |
| circRACGAP1 | _ | ceRNA | miR-144-5p/CDKL1 | Gefitinib | NSCLC | Oncogene | Up | [92] |
| circ_0009910 | Promote | ceRNA | miR-34a-5p/ULK1 | Imatinib | CML | Oncogene | Up | [93] |
| circCPM | Promote | ceRNA | miR-21-3p/PRKAA2 | 5-FU | GC | Oncogene | Up | [103] |
| circ_0035483 | Promote | ceRNA | miR-335/cyclinB1 | Gemcitabine | RCC | Oncogene | Up | [104] |
| circPARD3 | Suppress | ceRNA | miR-145-5p/PRKC1 | Cisplatin | LSCC | Oncogene | Up | [88] |
LSCC laryngeal squamous cell carcinoma, GC gastric cancer, NSCLC non-small cell lung cancer, CML chronic myeloid leukemia, RCC renal clear cell carcinoma.
The potential of autophagy-associated circRNAs in cancer treatment
With the increasing understanding of the regulatory roles played by circRNAs in autophagy and cancer development, the potential of circRNAs in tumor therapy is attracting more and more attention. The expression patterns of circRNAs have been shown that be associated with the clinicopathological characteristics of cancer. circHIPK3 plays an important role in regulating autophagy in NSCLC [59]. circHIPK3 and its linear partner (linHIPK3) exert the opposite regulatory effects on autophagy.The ratio between circHIPK3 and linHIPK3 (the C:L ratio) reflects the level of autophagy, with a low C:L ratio inducing autophagy in NSCLC. Additionally, a high C:L ratio indicates poor survival in patients with advanced-stage NSCLC, therefore it was suggested that the ratio can be used as a prognostic factor in NSCLC. Moreover, circMUC16 was reported to promote autophagy in EOC [55]. circMUC16 is upregulated in EOC tissues and the expression level of this circRNA is closely related to EOC development, both in terms of stage and grade. Therefore, circRNAs exhibit potential as novel targets for cancer diagnosis and therapy.
Some circRNAs are detectable in body fluids, such as peripheral blood, saliva, and urine, providing the possibility that they could be used as non-invasive diagnostic and prognostic biomarkers for numerous human diseases, including cancers [94]. Thus, the circRNAs enriched in exosomes have received more and more attention. Exosomes are tiny membrane vesicles circulating in body fluids, which are secreted by various types of cells, such as cancer cells [95]. They contain various substances, including circRNAs, other nucleic acids, lipids, and proteins, and are associated with intercellular communication and the formation of the tumor microenvironment and premetastatic niches [94, 96, 97]. Exosomes carry circRNAs to target cells and, at the same time, increase their stability [94]. Increasing numbers of cancer-derived exosomal circRNAs have been identified, and their expression level is related to tumor progression. As they are abundant, stable, easy to detect, and show specific expression patterns in cancer, these exosomal circRNAs are considered as novel potential biomarkers of cancer diagnosis and prognosis. Rao et al. compared circRNA levels in plasma exosomes between GC patients and healthy donors to identify the expression profile of exosomal circRNAs in GC [98]. They found 620 upregulated and 440 downregulated exosomal circRNAs, which may participate in GC development. Additionally, 1195 and 1147 dysregulated exosome-derived circRNAs were identified in localized and metastatic BCa, respectively [99]. Compared with patients with localized BCa, 480 exosome-derived circRNAs were dysregulated in metastatic BCa patients, indicating their role in BCa metastasis. Moreover, circSHKBP1 is upregulated in cancer tissues and serum of GC patients [100]. Its increased expression is linked to poor survival and advanced TNM stage. The exosomal circSHKBP1 enhances GC development via the miR-582-3p/HUR/VEGF axis and its level significantly decreases following gastrectomy. Therefore, exosomal circSHKBP1 might be a potential biomarker for GC. The enrichment of circSATB2 was found in serum exosomes from NSCLC patients and is linked to metastasis [101]. This exosomal circRNA, which can be detected with high sensitivity and specificity, also exhibit potential as a diagnostic biomarker for NSCLC. The expression patterns of autophagy-associated circRNAs in cancer-derived exosomes and their correlation with clinicopathological characteristics of cancers should be investigated further. The regulatory roles and the molecular mechanisms of these exosomal circRNAs during tumor development also need to be further elucidated.
Chemotherapy is primarily used for the treatment of metastatic cancers. However, that multidrug resistance is usually developed in patients renders cancer treatment more difficult. Autophagy-associated circRNAs play vital roles in chemoresistance development; therefore, these circRNAs can be targeted to enhance the sensitivity of cancer cells to anti-cancer reagents. The drug 5-fluorouracil (5-FU) is frequently used for the treatment of advanced GC [102]. Fang et al. reported that circCPM is upregulated in GC cell lines and tissues with resistance to 5-FU [103]. circCPM promotes PRKAA2-mediated autophagy by sponging miR-21-3p and then enhances 5-FU resistance of GC cells. Therefore, targeting circCPM may represent a novel therapeutic strategy to reverse 5-FU resistance in GC. Hsa_circ_0035483 promotes gemcitabine-induced autophagy and enhances gemcitabine resistance in renal clear cell carcinoma (RCC) via sponging has_miR_335 [104]. circ_0035483 knockdown enhances the gemcitabine sensitivity, suggesting its potential as a novel target of treatment to reverse chemoresistance in RCC.
Conclusion
circRNAs is implicated in the regulation of autophagy, via multiple signaling pathways. circRNA-mediated autophagy plays vital roles in multiple aspects of tumor progression, and exhibits both pro- and anti-tumor effects, in a context-dependent manner. Therefore, an understanding of the mechanisms by which circRNA-mediated autophagy affects tumorigenesis and cancer progression is particularly important for the development of novel circRNA-based cancer therapeutic strategies. Furtherly, circRNAs also participate in the chemoresistance of cancer cells through the regulation of autophagy. The combination of traditional chemotherapy and new therapeutic strategies targeting autophagy-associated circRNAs may lead to more effective treatments for cancer. Additionally, some circRNAs enriched in exosomes are stable and detectable in body fluids, while the expression levels of circRNAs are linked to clinicopathological characteristics of cancers. Therefore, circRNAs are also considered to represent novel and potentially useful diagnostic and prognostic biomarkers of cancer.
Author contributions
YY wrote the manuscript. XZ and XF drew the figures. YP and ZJ revised the manuscript. All authors have approved the final manuscript.
Funding
This research was supported by: National Natural Science Foundation of China (82173290, 82172946, 81772592, 31601028, 81871969); Medical science and technology research foundation of Guangdong Province (A2019211, A2018170); Shenzhen Basic Research Fund (JCYJ20190808163801777); Shenzhen University Talent program (000324), High Quality University Construction 2nd phase (860-00000210); National Key Research and Development Program of China (2016YFB0201305); Youth talent support program in medical center Shenzhen University.
Data availability
All the data used to support the findings of this study are available in the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yin Peng, Email: ypeng@szu.edu.cn.
Zhe Jin, Email: zhejin@szu.edu.cn.
References
- 1.Kos A, Dijkema R, Arnberg AC, Vandermeide PH, Schellekens H. The Hepatitis Delta (Delta) virus possesses a circular RNA. Nature. 1986;323:558–60. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 2.Ge JS, Wang J, Xiong F, Jiang XJ, Zhu KJ, Wang YA, et al. Epstein-Barr virus-encoded circular RNA CircBART2.2 promotes immune escape of nasopharyngeal carcinoma by regulating PD-L1. Cancer Res. 2021;81:5074–88. doi: 10.1158/0008-5472.CAN-20-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–85. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 4.Gokool A, Anwar F, Voineagu I. The landscape of circular RNA expression in the human brain. Biol Psychiat. 2020;87:294–304. doi: 10.1016/j.biopsych.2019.07.029. [DOI] [PubMed] [Google Scholar]
- 5.Huang R, Zhang Y, Han B, Bai Y, Zhou R, Gan G, et al. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy. 2017;13:1722–41. doi: 10.1080/15548627.2017.1356975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su H, Zheng W, Pan J, Lv X, Xin S, Xu T. Circular RNA circSamd4a regulates antiviral immunity in teleost fish by upregulating STING through sponging miR-29a-3p. J Immunol. 2021;207:2770–84. doi: 10.4049/jimmunol.2100469. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q, et al. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9:588–607. doi: 10.7150/thno.29678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M, et al. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun. 2019;10:4317. doi: 10.1038/s41467-019-11777-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu F, Han B, Wu S, Yang L, Leng S, Li M, et al. Circular RNA TLK1 aggravates neuronal injury and neurological deficits after ischemic stroke via miR-335-3p/TIPARP. J Neurosci. 2019;39:7369–93. doi: 10.1523/JNEUROSCI.0299-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiao K, Walsh LJ, Ivanovski S, Han P (2021) The emerging regulatory role of circular rnas in periodontal tissues and cells. Int J Mol Sci. 2021;22:4636. [DOI] [PMC free article] [PubMed]
- 11.Zhang F, Zhang R, Zhang X, Wu Y, Li X, Zhang S, et al. Comprehensive analysis of circRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of atherosclerosis in rabbits. Aging (Albany NY) 2018;10:2266–83. doi: 10.18632/aging.101541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6:319–36. doi: 10.1016/j.trecan.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 13.Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539–55. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829–42. doi: 10.1038/s41388-018-0369-y. [DOI] [PubMed] [Google Scholar]
- 16.Peng Y, Xu Y, Zhang X, Deng S, Yuan Y, Luo X, et al. A novel protein AXIN1-295aa encoded by circAXIN1 activates the Wnt/beta-catenin signaling pathway to promote gastric cancer progression. Mol Cancer. 2021;20:158. doi: 10.1186/s12943-021-01457-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang W, Fu K, Sun H, Rong D, Wang H, Cao H. CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer. 2018;17:137. doi: 10.1186/s12943-018-0888-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang Y, Ouyang X, Zhang F, Wang K, Lin Y, Sun B, et al. Circular RNAs expression profiles in human gastric cancer. Sci Rep. 2017;7:9060. doi: 10.1038/s41598-017-09076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sand M, Bechara FG, Sand D, Gambichler T, Hahn SA, Bromba M, et al. Circular RNA expression in basal cell carcinoma. Epigenomics. 2016;8:619–32. doi: 10.2217/epi-2015-0019. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Li Y, Li L. Circular RNA hsa_Circ_0005795 mediates cell proliferation of cutaneous basal cell carcinoma via sponging miR-1231. Arch Dermatol Res. 2021;313:773–82. doi: 10.1007/s00403-020-02174-y. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. 2019;18:119. doi: 10.1186/s12943-019-1046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han D, Li J, Wang H, Su X, Hou J, Gu Y, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–64. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 23.Yang BL, Liu GQ, Li P, Li XH. Circular RNA CUL2 regulates the development of colorectal cancer by modulating apoptosis and autophagy via miR-208a-3p/PPP6C. Aging (Albany NY) 2022;14:497–508. doi: 10.18632/aging.203827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong X, Liu N, Liang Y, He Q, Yang X, Lei Y, et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol Cancer. 2020;19:33. doi: 10.1186/s12943-020-01149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cen J, Liang Y, Huang Y, Pan Y, Shu G, Zheng Z, et al. Circular RNA circSDHC serves as a sponge for miR-127-3p to promote the proliferation and metastasis of renal cell carcinoma via the CDKN3/E2F1 axis. Mol Cancer. 2021;20:19. doi: 10.1186/s12943-021-01314-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu Y, Wang Y, He L, Zhang J, Zhu X, Liu N, et al. Circular RNA circIPO11 drives self-renewal of liver cancer initiating cells via Hedgehog signaling. Mol Cancer. 2021;20:132. doi: 10.1186/s12943-021-01435-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei CY, Zhu MX, Lu NH, Liu JQ, Yang YW, Zhang Y, et al. Circular RNA circ_0020710 drives tumor progression and immune evasion by regulating the miR-370-3p/CXCL12 axis in melanoma. Mol Cancer. 2020;19:84. doi: 10.1186/s12943-020-01191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Y, Leng K, Yao Y, Kang P, Liao G, Han Y, et al. A circular RNA, cholangiocarcinoma-associated circular RNA 1, contributes to cholangiocarcinoma progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology. 2021;73:1419–35. doi: 10.1002/hep.31493. [DOI] [PubMed] [Google Scholar]
- 29.Necula L, Matei L, Dragu D, Neagu AI, Mambet C, Nedeianu S, et al. Recent advances in gastric cancer early diagnosis. World J Gastroenterol. 2019;25:2029–44. doi: 10.3748/wjg.v25.i17.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filomeni G, De Zio D, Cecconi F. Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ. 2015;22:377–88. doi: 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–42. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janji B, Berchem G, Chouaib S. Targeting autophagy in the tumor microenvironment: new challenges and opportunities for regulating tumor immunity. Front Immunol. 2018;9:887. [DOI] [PMC free article] [PubMed]
- 34.Yang T, Shen P, Chen Q, Wu P, Yuan H, Ge W, et al. FUS-induced circRHOBTB3 facilitates cell proliferation via miR-600/NACC1 mediated autophagy response in pancreatic ductal adenocarcinoma. J Exp Clin Cancer Res. 2021;40:261. doi: 10.1186/s13046-021-02063-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Zhang X, Cao J, Xu P, Chen Z, Wang S, et al. Circular RNA UBE2Q2 promotes malignant progression of gastric cancer by regulating signal transducer and activator of transcription 3-mediated autophagy and glycolysis. Cell Death Dis. 2021;12:910. doi: 10.1038/s41419-021-04216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 37.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–47. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–34.. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. 2017;67:214–27 e7. doi: 10.1016/j.molcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 40.He J, Chu Z, Lai W, Lan Q, Zeng Y, Lu D, et al. Circular RNA circHERC4 as a novel oncogenic driver to promote tumor metastasis via the miR-556-5p/CTBP2/E-cadherin axis in colorectal cancer. J Hematol Oncol. 2021;14:194. doi: 10.1186/s13045-021-01210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li S, Liu F, Zheng K, Wang W, Qiu E, Pei Y, et al. CircDOCK1 promotes the tumorigenesis and cisplatin resistance of osteogenic sarcoma via the miR-339-3p/IGF1R axis. Mol Cancer. 2021;20:161. doi: 10.1186/s12943-021-01453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jarlstad Olesen MT, L SK. Circular RNAs as microRNA sponges: evidence and controversies. Essays Biochem. 2021;65:685–96. doi: 10.1042/EBC20200060. [DOI] [PubMed] [Google Scholar]
- 43.Wang X, Chen M, Fang L. hsa_circ_0068631 promotes breast cancer progression through c-Myc by binding to EIF4A3. Mol Ther Nucleic Acids. 2021;26:122–34. doi: 10.1016/j.omtn.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X, Liu Y, Liu Z, Lin C, Meng F, Xu L, et al. CircMYH9 drives colorectal cancer growth by regulating serine metabolism and redox homeostasis in a p53-dependent manner. Mol Cancer. 2021;20:114. doi: 10.1186/s12943-021-01412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu T, Ran L, Zhao H, Yin P, Li W, Lin J, et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol Ther Nucleic Acids. 2021;26:649–64. doi: 10.1016/j.omtn.2021.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma X, Chen H, Li L, Yang F, Wu C, Tao K. CircGSK3B promotes RORA expression and suppresses gastric cancer progression through the prevention of EZH2 trans-inhibition. J Exp Clin Cancer Res. 2021;40:330. doi: 10.1186/s13046-021-02136-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie F, Huang C, Liu F, Zhang H, Xiao X, Sun J, et al. CircPTPRA blocks the recognition of RNA N(6)-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Mol Cancer. 2021;20:68. doi: 10.1186/s12943-021-01359-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu J, Ji L, Liang Y, Wan Z, Zheng W, Song X, et al. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. 2020;5:298. doi: 10.1038/s41392-020-00375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen S, Yang Y, Shen P, Ma J, Fang B, Wang Q, et al. circPDE4B prevents articular cartilage degeneration and promotes repair by acting as a scaffold for RIC8A and MID1. Ann Rheum Dis. 2021;80:1209–19. doi: 10.1136/annrheumdis-2021-219969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu Z, Gu S, Wu K, Li L, Dong C, Wang W, et al. CircRNA-DOPEY2 enhances the chemosensitivity of esophageal cancer cells by inhibiting CPEB4-mediated Mcl-1 translation. J Exp Clin Cancer Res. 2021;40:361. doi: 10.1186/s13046-021-02149-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Chen B, Zhao J, Li Q, Chen S, Guo T, et al. HNRNPL circularizes ARHGAP35 to produce an oncogenic protein. Adv Sci (Weinh) 2021;8:2001701. doi: 10.1002/advs.202001701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Jiang J, Zhang J, Shen H, Wang M, Guo Z, et al. CircDIDO1 inhibits gastric cancer progression by encoding a novel DIDO1-529aa protein and regulating PRDX2 protein stability. Mol Cancer. 2021;20:101. doi: 10.1186/s12943-021-01390-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang GH, Ling Y, Mehrpour M, Saw PE, Liu ZH, Tan WG, et al. Autophagy-associated circRNA circCDYL augments autophagy and promotes breast cancer progression. Mol Cancer. 2020;19:65. [DOI] [PMC free article] [PubMed]
- 54.Wang HX, Sun GL, Xu PH, Lv JL, Zhang X, Zhang L, et al. Circular RNA TMEM87A promotes cell proliferation and metastasis of gastric cancer by elevating ULK1 via sponging miR-142-5p. J Gastroenterol. 2021;56:125–38. doi: 10.1007/s00535-020-01744-1. [DOI] [PubMed] [Google Scholar]
- 55.Gan XL, Zhu HT, Jiang XW, Obiegbusi SC, Yong M, Long XT, et al. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer. 2020;19:45. [DOI] [PMC free article] [PubMed]
- 56.Shen P, Yang T, Chen Q, Yuan H, Wu P, Cai B, et al. CircNEIL3 regulatory loop promotes pancreatic ductal adenocarcinoma progression via miRNA sponging and A-to-I RNA-editing. Mol Cancer. 2021;20:51. doi: 10.1186/s12943-021-01333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu H, Yuan HF, Xu D, Chen KJ, Tan N, Zheng QJ. Circular RNA circ_0000034 upregulates STX17 level to promote human retinoblastoma development via inhibiting miR-361-3p. Eur Rev Med Pharm. 2020;24:12080–92. doi: 10.26355/eurrev_202012_23997. [DOI] [PubMed] [Google Scholar]
- 58.You LK, Wang ZG, Li HS, Shou JW, Jing Z, Xie JS, et al. The role of STAT3 in autophagy. Autophagy. 2015;11:729–39. doi: 10.1080/15548627.2015.1017192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen XY, Mao R, Su WM, Yang X, Geng QQ, Guo CF, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPK alpha signaling in STK11 mutant lung cancer. Autophagy. 2020;16:659–71. doi: 10.1080/15548627.2019.1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang HJ, Liu YM, Wang DM, Xu YL, Dong RQ, Yang YX, et al. (2019) The upstream pathway of mtor-mediated autophagy in liver diseases. Cells. 2019;8:1597. [DOI] [PMC free article] [PubMed]
- 62.Gao L, Dou ZC, Ren WH, Li SM, Liang X, Zhi KQ. CircCDR1as upregulates autophagy under hypoxia to promote tumor cell survival via AKT/ERK(1/2)/mTOR signaling pathways in oral squamous cell carcinomas. Cell Death Dis. 2019;10:745. doi: 10.1038/s41419-019-1971-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shang J, Chen WM, Liu S, Wang ZH, Wei TN, Chen ZZ, et al. CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk Res. 2019;85:106198. doi: 10.1016/j.leukres.2019.106198. [DOI] [PubMed] [Google Scholar]
- 64.Levine B, Abrams J. p53: The Janus of autophagy? Nat Cell Biol. 2008;10:637–9. doi: 10.1038/ncb0608-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mrakovcic M, Frohlich LF. p53-mediated molecular control of autophagy in tumor cells. Biomolecules. 2018;8:14. [DOI] [PMC free article] [PubMed]
- 66.Maiuri MC, Tasdemir E, Criollo A, Morselli E, Vicencio JM, Carnuccio R, et al. Control of autophagy by oncogenes and tumor suppressor genes. Cell Death Differ. 2009;16:87–93. doi: 10.1038/cdd.2008.131. [DOI] [PubMed] [Google Scholar]
- 67.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–60. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng B, Lu J, Li T, Meng Z, Liu M, Sun M, et al. 1,3-Dichloro-2-Propanol inhibits autophagy via P53/AMPK/mTOR pathway in HepG2 cells. Food Chem Toxicol. 2018;122:143–50. doi: 10.1016/j.fct.2018.10.030. [DOI] [PubMed] [Google Scholar]
- 69.Zhu J, Ao H, Liu M, Cao K, Ma J. UBE2T promotes autophagy via the p53/AMPK/mTOR signaling pathway in lung adenocarcinoma. J Transl Med. 2021;19:374. doi: 10.1186/s12967-021-03056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crighton D, Wilkinson S, Ryan KM. DRAM links autophagy to p53 and programmed cell death. Autophagy. 2007;3:72–4. doi: 10.4161/auto.3438. [DOI] [PubMed] [Google Scholar]
- 71.Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–34. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 72.Cheng ZY. The FoxO-autophagy axis in health and disease. Trends Endocrin Met. 2019;30:658–71. doi: 10.1016/j.tem.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 73.Fitzwalter BE, Thorburn A. FOXO3 links autophagy to apoptosis. Autophagy. 2018;14:1467–8. doi: 10.1080/15548627.2018.1475819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen F, Guo L, Di J, Li M, Dong D, Pei D. Circular RNA ubiquitin-associated protein 2 enhances autophagy and promotes colorectal cancer progression and metastasis via miR-582-5p/FOXO1 signaling. J Genet Genomics. 2021;48:1091–1103. [DOI] [PubMed]
- 75.Jiang C, Jiang Z, Zhang X. Circular RNA circMRPS35 regulates progression and autophagy in osteosarcoma cells by recruiting KAT6B to govern FOXO3. Anticancer Drugs. 2022;33:607–13. doi: 10.1097/CAD.0000000000001276. [DOI] [PubMed] [Google Scholar]
- 76.Peeters JGC, Picavet LW, Coenen S, Mauthe M, Vervoort SJ, Mocholi E, et al. Transcriptional and epigenetic profiling of nutrient-deprived cells to identify novel regulators of autophagy. Autophagy. 2019;15:98–112. doi: 10.1080/15548627.2018.1509608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu J, Zhong C, Luo J, Shu F, Lv D, Liu Z, et al. HnRNP-L-regulated circCSPP1/miR-520h/EGR1 axis modulates autophagy and promotes progression in prostate cancer. Mol Ther Nucleic Acids. 2021;26:927–44. doi: 10.1016/j.omtn.2021.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Takeda T, Komatsu M, Chiwaki F, Komatsuzaki R, Nakamura K, Tsuji K, et al. Upregulation of IGF2R evades lysosomal dysfunction-induced apoptosis of cervical cancer cells via transport of cathepsins. Cell Death Dis. 2019;10:876. doi: 10.1038/s41419-019-2117-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Z, Mou Z, Xu C, Wu S, Dai X, Chen X, et al. Autophagy-associated circular RNA hsa_circ_0007813 modulates human bladder cancer progression via hsa-miR-361-3p/IGF2R regulation. Cell Death Dis. 2021;12:778. doi: 10.1038/s41419-021-04053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun L, Ma Y, Zhang Z, Li X, Chen Y, Liu G, et al. ROCK2 regulates autophagy in the hippocampus of rats after subarachnoid hemorrhage. Neuroreport. 2018;29:1571–7. doi: 10.1097/WNR.0000000000001154. [DOI] [PubMed] [Google Scholar]
- 81.Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X, et al. circCUL2 regulates gastric cancer malignant transformation and cisplatin resistance by modulating autophagy activation via miR-142-3p/ROCK2. Mol Cancer. 2020;19:156. doi: 10.1186/s12943-020-01270-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bae SY, Byun S, Bae SH, Min DS, Woo HA, Lee K. TPT1 (tumor protein, translationally-controlled 1) negatively regulates autophagy through the BECN1 interactome and an MTORC1-mediated pathway. Autophagy. 2017;13:820–33. doi: 10.1080/15548627.2017.1287650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang N, Zhang X, Xu W, Zhang X, Mu Z. CircRNA_103948 inhibits autophagy in colorectal cancer in a ceRNA manner. Ann N. Y Acad Sci. 2021;1503:88–101. doi: 10.1111/nyas.14679. [DOI] [PubMed] [Google Scholar]
- 84.Tang Q, Chen Z, Zhao L, Xu H. Circular RNA hsa_circ_0000515 acts as a miR-326 sponge to promote cervical cancer progression through up-regulation of ELK1. Aging (Albany NY) 2019;11:9982–99. doi: 10.18632/aging.102356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharm. 2014;740:364–78. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tchounwou PB, Dasari S, Noubissi FK, Ray P, Kumar S. Advances in our understanding of the molecular mechanisms of action of cisplatin in cancer therapy. J Exp Pharm. 2021;13:303–28. doi: 10.2147/JEP.S267383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Galluzzi L, Senovilla L, Vitale I, Michels J, Martins I, Kepp O, et al. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31:1869–83. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 88.Gao W, Guo H, Niu M, Zheng X, Zhang Y, Xue X, et al. circPARD3 drives malignant progression and chemoresistance of laryngeal squamous cell carcinoma by inhibiting autophagy through the PRKCI-Akt-mTOR pathway. Mol Cancer. 2020;19:166. doi: 10.1186/s12943-020-01279-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhong Y, Lin H, Li Q, Liu C, Shen J. CircRNA_100565 contributes to cisplatin resistance of NSCLC cells by regulating proliferation, apoptosis and autophagy via miR-337-3p/ADAM28 axis. Cancer Biomark. 2021;30:261–73. doi: 10.3233/CBM-201705. [DOI] [PubMed] [Google Scholar]
- 90.Fathi Maroufi N, Rashidi MR, Vahedian V, Akbarzadeh M, Fattahi A, Nouri M. Therapeutic potentials of Apatinib in cancer treatment: Possible mechanisms and clinical relevance. Life Sci. 2020;241:117106. doi: 10.1016/j.lfs.2019.117106. [DOI] [PubMed] [Google Scholar]
- 91.Ma L, Wang Z, Xie M, Quan Y, Zhu W, Yang F, et al. Silencing of circRACGAP1 sensitizes gastric cancer cells to apatinib via modulating autophagy by targeting miR-3657 and ATG7. Cell Death Dis. 2020;11:169. doi: 10.1038/s41419-020-2352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lu M, Xiong H, Xia ZK, Liu B, Wu F, Zhang HX, et al. circRACGAP1 promotes non-small cell lung cancer proliferation by regulating miR-144-5p/CDKL1 signaling pathway. Cancer Gene Ther. 2021;28:197–211. doi: 10.1038/s41417-020-00209-0. [DOI] [PubMed] [Google Scholar]
- 93.Cao HX, Miao CF, Sang LN, Huang YM, Zhang R, Sun L, et al. Circ_0009910 promotes imatinib resistance through ULK1-induced autophagy by sponging miR-34a-5p in chronic myeloid leukemia. Life Sci. 2020;243:117255. doi: 10.1016/j.lfs.2020.117255. [DOI] [PubMed] [Google Scholar]
- 94.Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116. doi: 10.1186/s12943-019-1041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–4. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lobb RJ, Lima LG, Moller A. Exosomes: Key mediators of metastasis and pre-metastatic niche formation. Semin Cell Dev Biol. 2017;67:3–10. doi: 10.1016/j.semcdb.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 97.Dai J, Su Y, Zhong S, Cong L, Liu B, Yang J, et al. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct Target Ther. 2020;5:145. doi: 10.1038/s41392-020-00261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rao M, Zhu Y, Qi L, Hu F, Gao P. Circular RNA profiling in plasma exosomes from patients with gastric cancer. Oncol Lett. 2020;20:2199–208. doi: 10.3892/ol.2020.11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang J, Zhang Q, Zhou S, Xu H, Wang D, Feng J, et al. Circular RNA expression in exosomes derived from breast cancer cells and patients. Epigenomics. 2019;11:411–21. doi: 10.2217/epi-2018-0111. [DOI] [PubMed] [Google Scholar]
- 100.Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19:112. doi: 10.1186/s12943-020-01208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang N, Nan A, Chen L, Li X, Jia Y, Qiu M, et al. Circular RNA circSATB2 promotes progression of non-small cell lung cancer cells. Mol Cancer. 2020;19:101. doi: 10.1186/s12943-020-01221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Machover D, Goldschmidt E, Chollet P, Metzger G, Zittoun J, Benavides M, et al. Treatment of advanced colorectal and gastric adenocarcinomas with 5-fluorouracil and high-dose folinic acid. J. Clin. Oncol. 1986;4:193–8. [PubMed]
- 103.Fang L, Lv J, Xuan Z, Li B, Li Z, He Z, et al. Circular CPM promotes chemoresistance of gastric cancer via activating PRKAA2-mediated autophagy. Clin Transl Med. 2022;12:e708. doi: 10.1002/ctm2.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yan L, Liu G, Cao H, Zhang H, Shao F. Hsa_circ_0035483 sponges hsa-miR-335 to promote the gemcitabine-resistance of human renal cancer cells by autophagy regulation. Biochem Biophys Res Commun. 2019;519:172–8. doi: 10.1016/j.bbrc.2019.08.093. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data used to support the findings of this study are available in the paper.