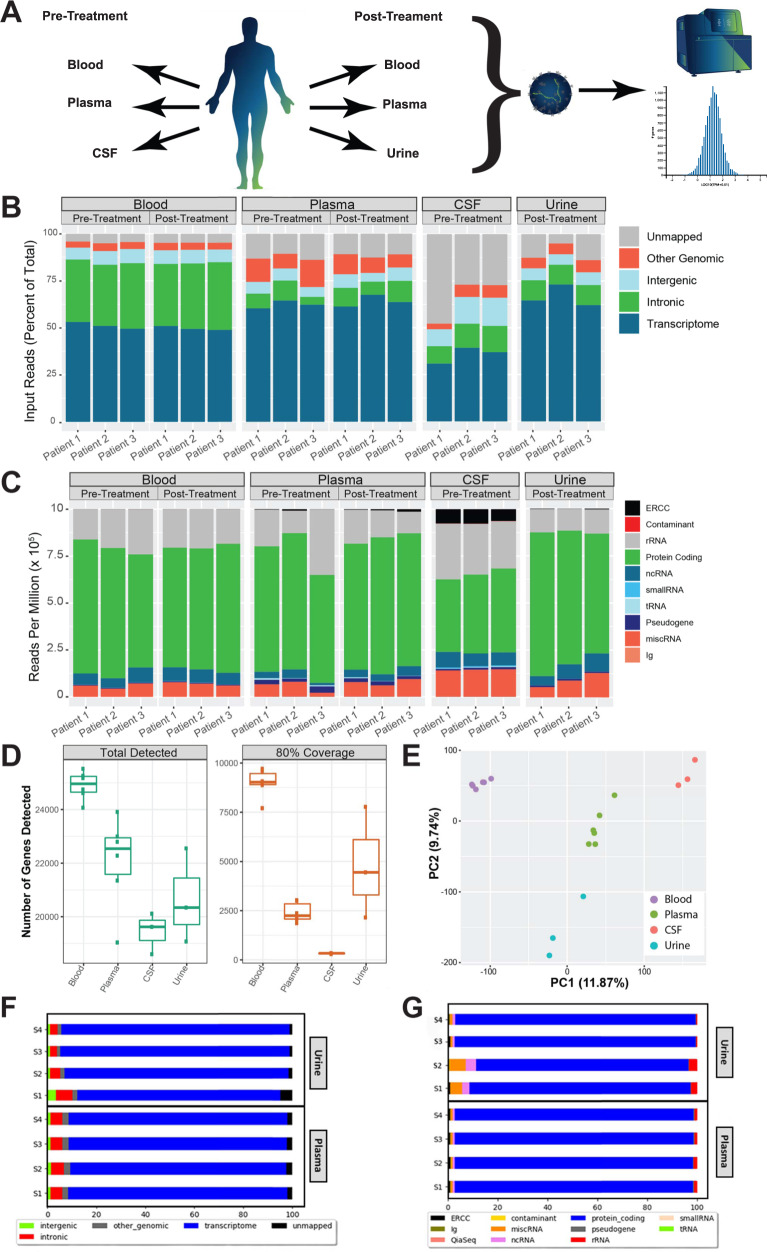
Fig. 1. High quality total RNA-Seq data on long RNAs extracted from extracellular vesicles from four different biofluids.
A A schematic summarizing experimental procedure. Whole blood, plasma, and CSF were collected from patients exhibiting high intercranial pressure (ICP) at their initial visit (“Pre-Treatment”) and whole blood, plasma, and urine at a follow-up visit (“Post-Treatment”) were subject to exoRNA isolation, RNA-Seq, and data analysis. B Stacked bar graphs showing the percentage of RNA-Seq reads mapping to transcriptome, intronic, intergenic, and other genomic regions by sample and biofluid type. C Stacked bar graphs showing the number of reads per million (RPM) mapping to various biotypes by sample and biofluid type. D Boxplots of the number of total genes detected and the number of genes covered at 80% or higher by biofluid. The center line represents the median, boxes represent ±1 interquartile range, and whiskers ±1.5 interquartile range. E Principal component analysis (PCA) of all samples showing clear separation of samples by biofluid type. F Exome capture performed on both plasma and urine RNA-Seq libraries resulted in increased proportion of reads mapped to the transcriptome when compared to total-RNA seq. G Exome capture performed on both plasma and urine RNA-Seq libraries resulted in increased proportion of reads mapping to protein coding genes and depletion of reads mapping to rRNA when compared with total RNA-Seq.