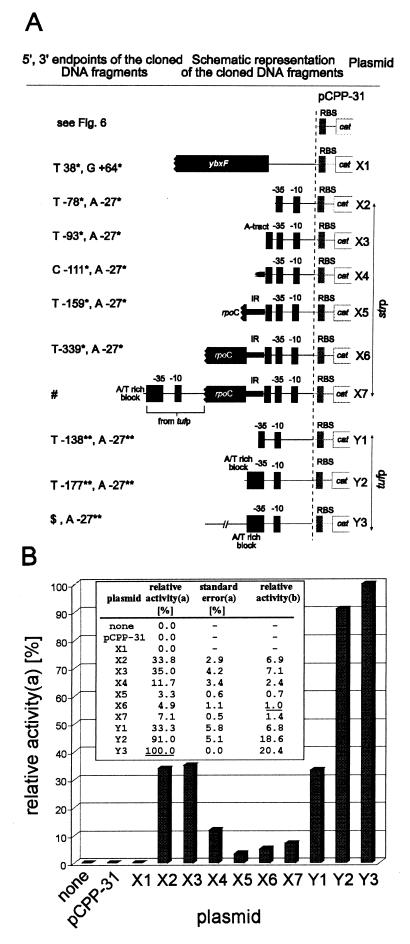
FIG. 8.
Schematic structure of pCPP-31 derivatives (A) and their transcriptional activity in CAT assays (B). (A) All DNA inserts were cloned into the EcoRI-PstI site of the polylinker upstream of the cat gene and its ribosome binding site (RBS) of pCPP-31. The picture is not drawn to scale, and the pCPP-31 part of the plasmid name is omitted. The ′X2-′X7 plasmids carry the strp core promoter sequence extended with its upstream DNA regions of increasing size as indicated. The ′Y1-′Y3 plasmids carry the tufp core promoter extended in the same way. ∗ and ∗∗, see Fig. 2 and 7. #, the DNA insert upstream of the strp core promoter is the same as in X6 extended for the tuf promoter region specified in ′Y2; $, the 5′ end of the insert is 499 bp upstream of the tuf gene translational start (for nucleotide sequence see EMBL database entry AJ249559). (B) CAT assay results. The background value of the negative control (plasmidless B. subtilis) was subtracted. The results are averages of at least two experiments done in duplicates, normalized to the same amounts of protein. a, the activity of the most active promoter region (pCPP-31.Y3) was taken as 100% (underlined); b, relative strengths of promoter regions compared to the activity of the strp promoter (pCPP-31.X6). Its activity was taken as 1 (underlined).