Figure 1.
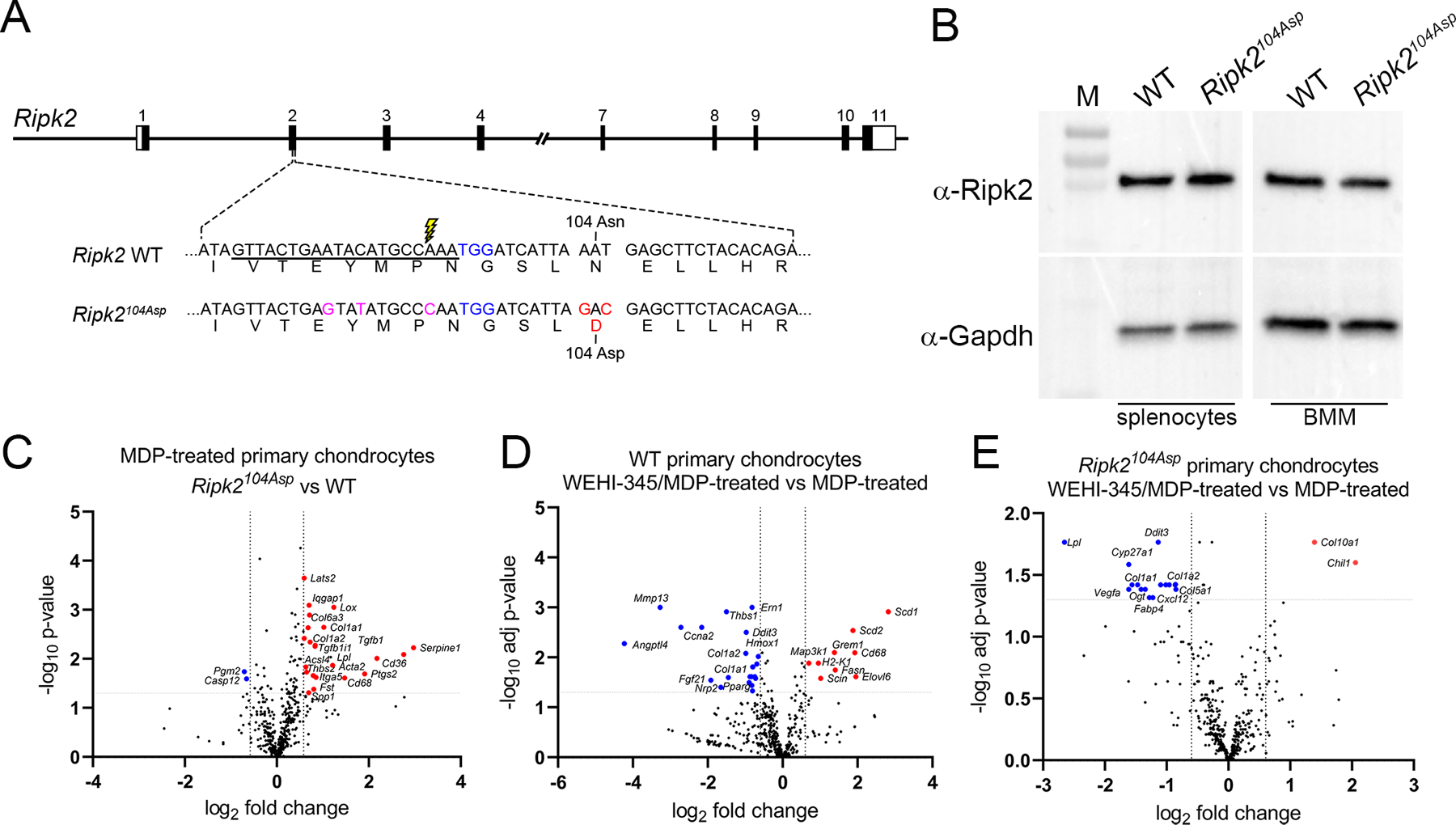
Generation and validation of the Ripk2104Asp mouse. (A) Schematic illustration of the mouse Ripk2 locus and detailed view of exon 2. CRISPR/Cas9-stimulated homology directed repair was used to edit sequences of C57Bl/6J mice (WT) encoding the Ripk2104Asn protein to generate an isogenic line that expressed the OA-associated Ripk2104Asp protein from the native locus. The guide RNA target sequence is underlined, the PAM site is highlighted in blue, and the Cas9 cleavage site is denoted with lightning bolt. An oligonucleotide donor was used as a template to create the mutations to generate Asp 104 (red) as well as silent mutations (magenta) to prevent targeting of the modified locus. (B) Immunoblot analysis indicates similar Ripk2 protein present in WT and Ripk2104Asp splenocytes or bone marrow derived macrophages (BMM). Gapdh is used as a loading control. M = protein mass standards in kDa. (C) A single copy of Ripk2104Asp is sufficient to alter the gene expression response of primary chondrocytes to MDP treatment. Volcano plots indicate genes significantly upregulated (red) or downregulated (blue) in MDP-treated Ripk2104Asp as compared to MDP-treated WT primary chondrocytes. (D and E) Increased gene expression in response to MDP stimulation is dependent on Ripk2 activity. D) WT or E) Ripk2104Asp primary chondrocytes were stimulated with MDP in the presence or absence of the Ripk2 inhibitor, WEHI-345. Volcano plots indicate genes significantly upregulated (red) or downregulated (blue) upon MDP-stimulation in the presence of the inhibitor.
