Abstract
Prokaryotic organisms frequently use riboswitches to quantify intracellular metabolite concentration via high-affinity metabolite receptors. Riboswitches possess a metabolite-sensing system that controls gene regulation in a cis-acting fashion at the initiation of transcriptional/translational level by binding with a specific metabolite and controlling various biochemical pathways. Riboswitch binds with flavin mononucleotide (FMN), a phosphorylated form of riboflavin and controls gene expression involved in riboflavin biosynthesis and transport pathway. The first step of the riboflavin biosynthesis pathway is initiated by the conversion of guanine nucleotide triphosphate (GTP), which is an intermediate of the purine biosynthesis pathway. An alternative pentose phosphate pathway of riboflavin biosynthesis includes the enzymatic conversion of ribulose-5-phosphate into 3, 4 dihydroxy-2-butanone-4-phosphates by DHBP synthase. The product of ribAB interferes with both GTP cyclohydrolase II as well as DHBP synthase activities, which catalyze the cleavage of GTP and converts DHBP Ribu5P in the initial steps of both riboflavin biosynthesis branches. Riboswitches are located in the 5′ untranslated region (5′ UTR) of messenger RNAs and contain an aptamer domain (highly conserved in sequence) where metabolite binding leads to a conformational change in an aptamer domain, which modulate the regulation of gene expression located on bacterial mRNA. In this review, we focus on how riboswitch regulates the riboflavin biosynthesis pathway in Bacillus subtilis and Lactobacillus plantarum.
Keywords: Lactobacillus, Riboswitch, Metabolite, Aptamer, Riboflavin, Gene regulation
Introduction
Riboswitches are highly conserved structural domains associated with mRNAs that recognize metabolites and participated in the regulation of gene expression (Saberi et al. 2016). These small mRNA elements possess binding affinity with different metabolites and subsequently harmonize transcription and translation with similar approaches resembling protein genetic factors (Scull et al. 2021). This appropriate metabolite binding system contains evolutionary sequences that might pre-date the emergence of the proteins in the living system (Micura and Höbartner 2020). Typically, riboswitches consist of a conserved metabolite binding domain (aptamer) and variable sequence region (expression platform) located in the 5′ UTR of mRNAs. The binding of the metabolite brings an allosteric change in the aptamer domain leading to an expression platform to modulate the expression of adjacent genes or operons (Jones and Ferré-D’Amaré 2017). Flavin mononucleotide (FMN) riboswitch is structurally more complex, located upstream of the rib operon and controls the expression of both riboflavin biosynthesis and transporter genes in certain bacteria (Table 1) (Lins et al. 2021). FMN binding domain/component of this riboswitch was originally entitled RFN element (Panchal and Brenk 2021). FMN riboswitch is located 5′ upstream of the ribDEATH operon in Bacillus subtilis binds with riboflavin 100-folds more tightly than FMN due to the absence of a phosphate group on riboflavin (Winkler and Breaker 2005).
Table 1.
Distributions of FMN riboswitch in different prokaryotic genera (Winkler and Breaker 2005)
| Name and functions of the riboswitch | Representative genera |
|---|---|
|
FMN riboswitch Functions: Control gene expression of riboflavin biosynthesis and transport |
Proteobacteria: α-proteobacteria (Mesorbizobium, Bradyrbizobium, Sinorbizobium, Brucella, Agrobacterium) β-proteobacteria (Nitrosomonas, Methylobacillus, Ralstonia) γ-proteobacteria (Escherichia, Salmonella, Klebsiella, Yersinia, Vibrio, Pasteurella, Pseudomonas, Xanthomonas) δ-proteobacteria (Geobacter) Deinococcales (Deinococcus) Bacillales (Bacillus, Listeria) Lactobacillales (Enterococcus) Clostridiales/Thermoanaerobacteriales (Clostridium, Thermoanaerobacter) Actinomycetales (Corynebacterium, Mycobacterium, Propionibacterium, Streptomyces) Cynobacteria (Anabaena, Thermosynecbococcus) Chlorobiales (Chlorobium) Bacteriodales (Bacteriodes) Thermotogales (Thermotoga) Spirobactales (Treponema, Leptospira) Archaea (Aeropyrum, Sulfolobus, Pyrococcus) |
Bold text indicates different classes of phylum “Proteobacteria”
In lactic acid bacteria (LAB), riboflavin biosynthesis genes are sequentially arranged and form a single ribGBAH operon or rib operon controlled by FMN riboswitch (Capozzi and Russo 2012). Modulation of gene expression by RFN element does not require any regulatory protein factors rather than the basal gene expression system of an organism. FMN Riboswitch can influence the generation of an intrinsic termination–hairpin loop that leads to prematurely terminating transcription or can favor forming RNA secondary structure which occludes ribosome binding (Chen et al. 2019). Generally, riboswitches stop the synthesis of enzymes or transporter when these are already available at a sufficient level. Under certain circumstances, such as increased production of metabolites, riboswitches can shift to the salvage pathway, which is involved in the degradation of excess metabolites (Sherlock et al. 2018). Certain riboswitches exhibit advanced mechanisms in gene expression systems, such as cooperative ligand binding (Mccown et al. 2017), self-cleavage and tandem aptamer arrangements (Tickner and Farzan 2021).
Genetic organization of the rib operon
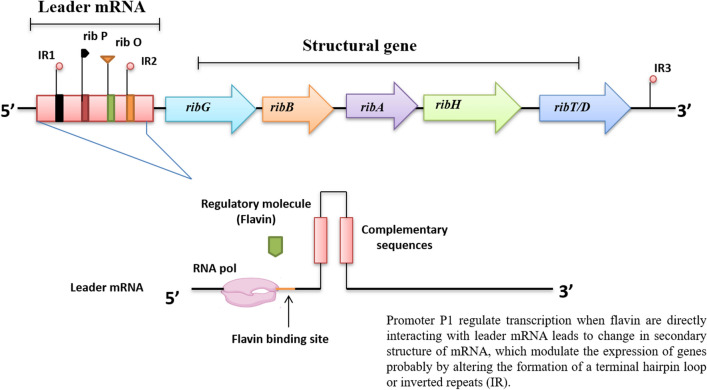
Riboflavin production in Bacillus subtilis is accomplished by the riboflavin biosynthesis operon i.e. rib operon which completely controls the riboflavin biosynthesis pathway from its initial precursor molecule GTP (Zhang et al. 2021). The genetic makeup of rib operon incorporates five non-overlapping structural genes and three regulatory elements represented as regulatory region ribO with promoter P1 and two internal supplementary promoters P2 and P3. P1 is the major promoter located in the 5′ upstream region and P2 and P3 are located in the distal region of the ribB and ribH respectively (Averianova et al. 2020). The major promoter P1 (TTGGGT-17bp-TATAAT) of the rib operon is recognized by S1 mapping (Mironov et al. 1994). Promoter P1 regulates transcription when flavin has directly interacted with leader mRNA leading to a change in the secondary structure of mRNA (Polaski et al. 2016), which modulates the expression of genes probably by altering the formation of the terminal hairpin loop. In this process, FMN riboswitch (RFN element), widely distributed in some prokaryotic genera (Table 1), is a transcript of leader mRNA that plays a very crucial role by providing a flavin binding site (Irastortza-Olaziregi and Amster-Choder 2021). The structural and functional role of internal supplementary promoters P2 and P3 is still unclear. Presumably, the P2 promoter is involved in the riboflavin biosynthesis mechanism (Wilt et al. 2020). Riboflavin precursors 5-amino-6-ribitylamino-pyrimidinedione and 6, 7 dimethyl-8-ribitylluzine are synthesized by both de novo and riboflavin biosynthesis pathways, respectively. However, the constitutive expression of ribA and ribH stabilize the riboflavin intermediates, such as pyrimidine and pteridine (Sklyarova et al. 2012). Promoter P3 is located upstream of the ribTD gene, the function of the T protein is not yet been reported. Inactivation of the TD gene does not originate riboflavin auxotrophy, but observed a significant decrease in riboflavin biosynthesis in producer organisms (Fuentes Flores et al. 2017).
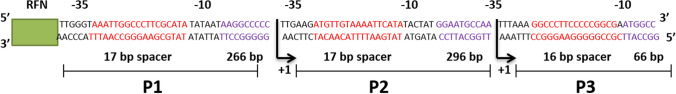
The transcription initiation site of the P2 promoter is located in the T base-rich region of the mRNA still; some subsidiary signals are also present both downstream and upstream of this region (Haberle and Stark 2018). It is proposed that the structural sequence of the P2 promoter is probably TTGAAG (-35 region) and TACTAT (-10 region) segregated by a 17 bp spacer (Shimada et al. 2014). A fragment of 296 base pairs is located between the transcription initiation site of the P2 promoter and the initiation codon of the ribA gene (Sklyarova et al. 2012). Based on previous studies the structural arrangement of the P3 promoter (Oliveira et al. 2017) is likely TTTAAA (-35 region) and TATAAT (-10 region) separated by 16 bp spacer and 66 bp sequence is present between the transcription initiation site of P3 promoter and initiation codon of ribT gene (Fig. 1).
Fig. 1.
The genetic organization of the rib operon in Bacillus subtilis
In Bacillus subtilis, riboflavin biosynthesis genes (rib gene cluster) are located on the SHgw chromosome. As previously reported from seven ORFs, only five are reported within the sequence, which constitutes genes ribG, ribB, ribA, ribH and ribTD. The mRNA of these five-rib genes is polycistronic in nature and consists of 4277 nucleotides (Thakur et al. 2017). Computer analysis of these seven ORFs revealed that the encoded proteins possess molecular mass 12,754 (ORF X), 39,305 (ORF 1), 23,481 (ORF 2), 44,121 (ORF 3), 16,286 (ORF 4), 14,574 (ORF 5) and 15,551 (ORF Y) Daltons where X and Y orientation are opposite to others. Each ORF is preceded by the Shine–Dalgarno sequence (SD). The ORF 2 region could be potentially translated from three nearby codons ATG (2299), GTG (2302) and ATG (2308) while the last codon ATG possesses the best SD sequence (Kil et al. 1992; Wang et al. 2021). In addition, a small ORF (ORF 0) is also reported which precedes and partially overlaps ORF 1 with a good SD sequence (Orr et al. 2021). The sequence of rib operon attribute three terminator–hairpin structures including IR sequences (IR 1, IR 2 and IR 3) with a T-rich fragment at the end of the 3' terminal. Most probably, the function of IR 1 and IR 2 is transcription termination from the downstream of the ORF X and ORF 5, respectively (Henkin et al. 1992; Di Salvo et al. 2019). The absence of a terminator-like-hairpin structure at the 3’ end of the ORF 1 along with ORF 4 and due to the shortness of IGR indicated that ORF 1 along with ORF 5 is co-transcribed. The function of IR 2 is rarely studied when compared with IR 1. It might be expected that IR 2 controls downstream gene expression (Lah et al. 2011). IR sequences are usually located at the 5' end of the transcriptional unit in Bacillus subtilis in some cases it is also observed that these are essential for gene regulation (Shaw and Fulco 1992; Helmann 2019).
Strength of promoters and interactions between ORFs and riboflavin biosynthesis genes
Corresponding regions of the promoters P 1, P 2 and P 3 is cloned into an integrative expression vector pDG268, which contains the LacZ reporter gene and then introduced into the chromosome of Bacillus subtilis strain RKH25 at amylase locus and other homologous variants (Cao et al. 2017). The strain RK25-C1 is not able to synthesize FMN due to a mutation in the ribC gene. The strains were observed at the transcription level with each promoter by evaluating β-galactosidase activity (Pedrolli et al. 2015b). Evaluating the β-galactosidase level in wild-type Bacillus subtilis strain RKH25 recommended that the P 2 promoter is 10–20 times lower than the major promoter of rib operon, P1 promoter whereas the P 3 promoter, increases the efficiency of the P 1 promoter by five times (Kil et al. 1992; Hoffmann et al. 2021). Comparisons of β-galactosidase activities under the control of promoters P1, P2 and P3 by evaluating the effect of flavins on the expression of all three promoters (Lara et al. 2017; Micura and Höbartner 2020). It is observed that only the P1 promoter increased (tenfold) activity. A 4.3 kb transcript is reported with a transcription start site upstream of ORF 1 and a termination site just downstream of ORF 5, covering all riboflavin biosynthesis genes. The translation product of ORF 4 is identical to lumazine synthase (ribH gene product) except for one amino acid of Bacillus subtilis (Lin et al. 2001). In Bacillus subtilis, the amino acid sequence of an enzyme riboflavin synthase encoded by the ribB gene, which is varying from the translated product of ORF 2 due to the absence of 13 C-terminal amino acids. The ribT and ribD genes are located in the ORF 5 region of the rib operon but in all other species, these genes might be genetically separable and possess different functions (Vitreschak et al. 2002). In the exponential growth phase of Bacillus subtilis, ORF Y transcript is not observed, which means ribY does not pertain to rib cluster. At the 5' end of the transcript, it is believed that a promoter-like element is present in Bacillus subtilis (Sudzinová et al. 2021), which is mostly recognized by RNA polymerase, as a result, rate of transcription is maximum at the late exponential growth phase. Inverted Repeats 3 (IR3) is located in the T-rich region from 47 bp downstream of ORF 5 at 3′end of polycistronic mRNA which influences the transcription termination in Bacillus subtilis (Sklyarova et al. 2012). Ultimately this extended fragment between transcription and translation initiation site incorporates small stretches of palindromic sequences that can form a hairpin loop structure which is possibly targeted transcriptional regulators involved in riboflavin biosynthesis (García-Angulo 2017).
Riboswitches (structural aspects)
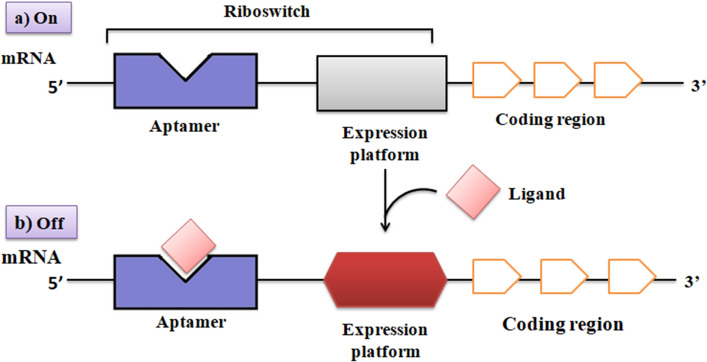
Riboswitches are small stretches that frequently originate in the 5′ UTR of the mRNA that strive its regulatory control over the transcribed mRNA in a cis-manner by binding with a small metabolite i.e. ligand (Ray et al. 2019). Usually, riboswitches consist of two recognizable functional domains. An aptamer domain recognized an effector molecule as a receptor and create a specific compact structure from three-dimensional to scaffold providing a ligand-binding pocket (Page et al. 2018). Like proteins, these RNA receptors can distinguish among the chemically different metabolites with high selectivity and are capable to elicit an appropriate regulatory response. A second domain is the ‘expression platform’ consisting of a regulatory switch that interferes with the transcriptional and translational machinery (Abduljalil 2018). Regulation is accomplished by the evolutionary conserved regulatory sequence of an element located in the overlapping region of these two domains (Cheng et al. 2021). The sequence is originally known as the ‘switching sequence’ that pairs directly with the folding of the mRNA into either mutually incompatible structure in the expression platform that acts as an on and off switch (Fig. 2) of the mRNA (Garst et al. 2011).
Fig. 2.
General organization of riboswitch RNAs in bacterial mRNAs. The binding of the ligand to the aptamer domain stabilizes an altered conformation of the expression platform that results in a change in the regulation of gene expression
This model of the riboswitch highlights two fundamental points regarding their regulatory mechanism. First, the formation of a binding pocket performs both high affinity and specificity with different effector molecules. The second is binding of the effector communicated with the expression platform to initiate regulatory outcome (Batey 2012). To understand this, knowledge is required of the potential of ligand-induced conformational changes in the binding pocket (aptamer) and co-transcriptional turn-up mechanism of the mRNA (Gong et al. 2017). Since the transcriptional attenuation mechanism is the prevalent approach of riboswitches regulation (Barrick and Breaker 2007), they quickly acquire the essential conformations to perform before running off RNA polymerase away from its 3'end (Mehdizadeh Aghdam et al. 2017). As previously reported that rigid coupling of the transcription and translation mechanism in prokaryotes may convey a similar transient obstruction on translation (Irastortza-Olaziregi and Amster-Choder 2021).
Riboswitches are fragments of RNAs organized as a receptor using numerous similar architectural principles recognized in other giant RNAs (Bou-Nader and Zhang 2020). In the purine riboswitch, the structural sequence of the aptamer domain consists of three evolutionary conserved helical fragments (P1, P2 and P3) (Fig. 3) that are existing in all ~ 500 determined sequences (Liberman et al. 2015). These helices are arranged in high-order structures of RNA by two principles (Holbrook 2008). First, they are arranged in sets of coaxial stacks inside of which at least two or more solitary helices are organized into an individual pseudo-continuous helix, illustrated through the P1 and P3 stack.
Fig. 3.
The structure of promoters P1, P2, and P3 of rib operon in Bacillus subtilis
Second, helices and stacks of helix accomplish to be organized into a parallel configuration. This type of structural arrangement is apparent in approximately all RNAs possessing three or more helices (Heinemann and Roske 2020). A remarkable exception to this arrangement is tRNA, inside of which two coaxial stacks are aligned perpendicularly through the synergy of the D-loops and T-loops. In addition, RNA helices are arranged in sequence motif that supports secondary structure segments: internal bulges and loops, terminal loops and multi-helix junctions (Hendrix et al. 2005; Bittrich et al. 2020). One of the unexceptional RNA motifs is the GA3 tetraloop, which contains the first guanosine nucleotide along with the three-adenosine nucleotides. This tetraloop represents a guanosine stack that is recognized by other RNA motifs including the tetraloop receptor (Wu et al. 2012), which was reported in the subgroup of di-cyclic guanosine monophosphate riboswitches (Sudarsan et al. 2008). Two helices are anchored together in a side-by-side adjustment throughout the interaction between loops and receptors that facilitate parallel packing. Variations were also observed in this terminal loop–internal loop in the structures of the thiamine pyrophosphate (TPP) and lysine riboswitches to execute the same structure consequences (Garst et al. 2011; Anderson et al. 2019), sarcin–ricin loops (Grundy et al. 2003), pseudoknots (McDaniel et al. 2005) and T-loops (Barrick and Breaker 2007). Some riboswitches possess multiple-helix junctions that contributed an important part of riboswitches while their structure and sequences vary, reflecting exclusive requirements for arranging the flanking helices or introducing active sites (Scull et al. 2021).
Recognition mechanisms of effector molecule by riboswitch
Properly folded structure of aptamer domain must succeed with two interlinked cascade processes: (1) Recognition of specific effector molecule and (2) communication with expression domain. The first objective needed that the RNA must be able to discriminate among similar compounds present in the cell. For instance, the purine riboswitches accomplish more than 10,000-fold strength to discriminate between guanine and adenine (Husser et al. 2021). Although lysine riboswitch accomplished over a 5000-fold strength of discrimination between lysine and ornithine. In addition, amino acids are discriminated by 5000-fold strength due to the presence of a single methylene group at their side chain (Irla et al. 2021). Based on their effector molecule the purine riboswitches family is divided into three classes, guanine/ frequent RNA building blocks that are observed in riboswitches, such as kink-turns (k-turns) (Schroeder et al. 2011), kissing-loop interactions hypoxanthine, adenine and 2′-deoxyguanosine (Polaski et al. 2016). The purine riboswitch family of aptamers is described by an ordinary conserved secondary and tertiary structure. In each class of this family, the effector molecule is held by a pocket constructed by the RNA three-way junction (Matyjasik and Batey 2019). The five-base triples model between the nucleotide defines the basic architecture of the binding pocket within the junction. At the central region, the base triple is located and involved in ligand binding and the other two-pyrimidine residues are involved in the recognition of a particular nucleobase (Espah Borujeni et al. 2016). Each functional group of the effector molecule is precisely recognized by H-bonding interactions, moderately justifying the capability of the RNA to attain high specificity for this particular compound (Weinberg et al. 2017). Especially, the nucleobase does not stack above or below nitrogenous bases, this feature is commonly found in the other riboswitches that recognize nucleobase-containing effector molecules (Stoddard et al. 2008). A base-triple model of two pyrimidine residues with the effector molecule attains the purine, specificity. The nucleotide located at the 74 positions participates in the Watson–Crick base pair with the ligand favor the RNA to distinguish between adenine and guanine/2’-deoxyguanosine (Balasubramaniyam et al. 2021). The position of this nucleotide is always exhibited in uridine and cytosine in adenine riboswitches and guanine/2′-deoxyguanosine riboswitches respectively. Similarly, the nucleotide located at position 51 forms a Watson–Crick base pairing with a ligand that is used to distinguish between nucleosides and nucleobases (adenine and guanine) (Balasubramaniyam et al. 2021). In adenine and guanine riboswitches the nucleotide 51 always acts as uridine whereas in 2′-deoxyguanosine riboswitches it acts as cytosine. This cytosine shifts in the close vicinity of the nucleotide 74 to open the binding pocket to recognize the presence of 2′-deoxyribose sugar moiety (Sherlock et al. 2018). In consequence, the small sequence might change to the aptamer structure and can create new riboswitches that deal with chemically distinct effector molecules.
Ligand binding induces conformational changes in the receptors
The effector molecules ensure that the aptamer domain must acquire a flexible "open" state that permits the effector molecule to enter into the binding pocket, as a result, the conformational change in the region of the effector-binding site. This phenomenon known as an induced-fit-binding mechanism is a very ordinary attribute of RNA binding processes, acting as in RNP structure organization (Kovach 2021). The conformational changes in the aptamer domain of riboswitches are responsible to control the regulatory switch in the expression platform which instructs the transcription and translation mechanisms (Sinumvayo et al. 2018). The Nuclear Magnetic Resonance (NMR) studies of purine aptamers pbuE and xpt-pbuX reported that in the absence of effector molecules, the nucleotides are completely disordered in a specific region called three-way junction (Matyjasik and Batey 2019) even at comprehensive temperatures the peaks of corresponding nucleotides are absent (Noeske et al. 2007). These regions are continuously observed as the same at the intensity of reactivity determined by SHAPE chemistry (Hurst et al. 2018) and the in-line probing method (Mandal et al. 2003). The data obtained from both NMR and chemical probing methods indicated that the regions J1/2 and J2/3 of the aptamer domain are disordered due to the absence of effectors molecule (Miao and Westhof 2017), perhaps these elements make a flexible lid that covers the docked ligand. In lysine riboswitch, ligand-induced folding of the aptamer domain is identical to the purine riboswitch. The approach of small-angle X-ray scattering (SAXS) proposed the global conformation of the ligand-binding pocket in the solution (Drogalis and Batey 2020), indicating that lysine aptamer is predetermined by Mg++. Although on the N-methyl isatoic anhydride (NMIA) chemical probing data exhibit a subset of nucleotides of five-way junction present in a disorder manner under these disproportionate conditions or absence of lysine (Garst et al. 2008). In the presence of an appropriate concentration of lysine, the subset of nucleotides is non-reactive to NMIA, demonstrating that lysine has participated in the local organization of the five-way junction. Interestingly, the fragments of RNA crystallize adequately in the lacking conditions of lysine (Chillón and Marcia 2020), enabling the free-state structure of aptamer for further characterization. Even though the crystal structure of the aptamer in both state free as well as bound is almost similar. This indicates that the aptamers behave like an ensemble of reciprocates structures in the solution and lysine restructures the other binding component of the RNAs. The aptamer can change its conformation and adopt a "bound-like" structure in the absence of an effector molecule so lysine is necessary to stabilize the crystallographically obtained structure (Novoseltseva et al. 2018).
Riboswitch genetic control mechanisms
The feedback repression mechanism of genes inside a cell is a dominant system to control genetic regulation, despite the several riboswitches involved in the gene activation by sensing the metabolite concentration (Bervoets and Charlier 2019). It is not caused by any limitations of the regulatory system or any deficiency in the RNA functions that have caused riboswitches to be involved in gene regulation. Somewhat, in bacteria, there is an incessant need to sense the excess concentration of metabolites and to stop the expression of particular genes involved in the biosynthesis or transport pathway (Machtel et al. 2021). But in a cell, it is less common to sense a similar compound and activates gene expression for that particular compound (Winkler and Breaker 2005). The riboswitches are predominantly used two basic mechanisms to control gene regulation i.e. control at transcription termination and control at the translation initiation level. Although these mechanisms can harness equal changes in the folding of the RNA structures contain Watson–Crick base pairing (Scull et al. 2021). The natural structure of aptamers is sub-sequentially reorganized upon metabolite binding (Hermann and Patel 2000). After binding of metabolite the aptamer converts into a stable form of the secondary and tertiary structure whereas, in the absence of metabolite, the aptamer reveals heterogeneous nature and is involved in the formation of alternative structure over the expression platform (Findeiß et al. 2017). This metabolite-binding-dependent folding of the aptamer can be exploited in many ways to regulate the expression of the proximate coding region of the mRNA (Ge and Marchisio 2021).
FMN riboswitch (RFN element)-mediated attenuation mechanisms of rib operon
Riboflavin biosynthesis metabolic pathway is extensively distributed in eubacteria. But especially in Spirochetes, Rickettsia and Mycoplasmas have neither rib operon nor RFN element (Vitreschak et al. 2002). The RFN element is an evolutionarily conserved region and is only located upstream of the riboflavin biosynthesis and transporter genes. The riboflavin biosynthesis genes form a single DEA/BTH operon (Pedrolli et al. 2015a) in several LAB and Bacillus/Clostridium groups except for Streptococcus pyogenes, Enterococcus faecalis and Listeria (Thakur et al. 2017). The absence of rib operon observed in some bacterial species is compensated by the presence of the riboflavin transporter YpaA gene (Vogl et al. 2007). In the group of proteobacteria, the redundancy of riboflavin biosynthesis genes is reported due to the presence of paralog genes of ribH, ribE and ribB/A (Fuentes Flores et al. 2017). Furthermore, in some bacteria, ribB/A genes are fused or present as additional genes. In proteobacteria, tightly RFN-regulated genes of the riboflavin biosynthesis pathway are ribB and ribH2. The ribH2 gene is a paralog to ribH gene and is located in the genome of some α-proteobacteria (Nouwen et al. 2021). The RFN elements are divided into two major groups based on the presence of two evolutionary conserved regions; AGCG and GTCAGCA, which exist inside the branching loops. The RFN element recognized FMN which influences the structural rearrangement of the mRNA (Winkler et al. 2002). In the interhelical regions of the RFN element certain nucleotides break spontaneously from their original position in the presence of FMN this suggested that these nucleotides interact with FMN and form a stable FMN–RNA complex, which controls the structural restraint to the mRNA (Ganser et al. 2020). It was reported that FMN regulates the riboflavin biosynthesis operon and transporter ypaA genes in Bacillus subtilis. Here we briefly illustrate the possible mechanism of FMN-mediated regulation of rib operon via RFN element (Averianova et al. 2020).
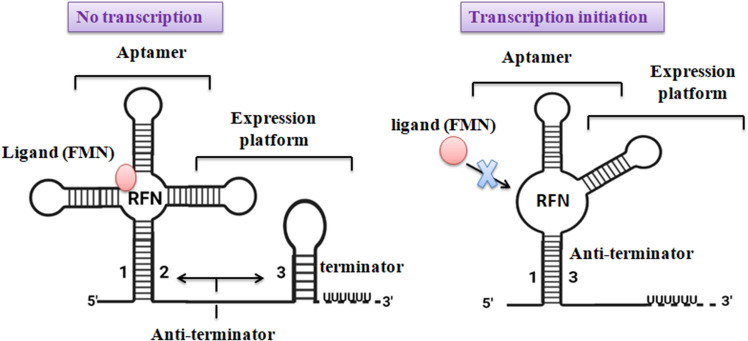
In the downstream of all RFN elements, either there is a possibility to form a hairpin structure that is succeeded by thymidine (terminators) or crisscrossing the translation initiation site of the first transcribing gene of the operon (sequesters) (Serganov et al. 2009; Gupta and Pal 2021). Additionally, the RFN elements efficiently form alternative structures, inside of which the base stem of RFN elements communicates to the regulatory hairpin (terminator/sequester). This trigger configures a structure substitute to the regulatory hairpin, involved in transcriptional and translational attenuation mechanisms (Wang et al. 2019). Hence, two different regulation mechanisms are reported, possible attenuation of transcription by anti-terminator and possible attenuation of translation via sequestering of the Shino–Dalgarno (SD) sequence (Kochhar and Paulus 2010). Gram-positive bacteria contain complementary sequences located between the RFN element and translation initiation site capable to form a hairpin loop which acts as a Terminator-like structure of RNA (Chełkowska-Pauszek et al. 2021).
Moreover, these complementary sequences are always involved in the formation of new stable secondary structures, as previously reported these structures act as anti-terminator (Hua et al. 2020). Excess concentration of FMN inside a cell leads to preventing the formation/configuration of an anti-terminator hairpin loop subsequently terminator is formed and transcription is initially terminated (Strobel et al. 2020). In the absence of FMN, the unstable structure of RFN is replaced by stable and anti-termination favorable conformation that allows the transcription continuously (Wilt et al. 2020) (Fig. 4). In some riboswitches, the interior of the expression platform is a helical element, which is varying in length and the region of the SD sequence. This helical element is called sequestering-helix because it has the capability of base pairing to the SD sequence region (Bhagdikar et al. 2020).
Fig. 4.
The predicted attenuation mechanism of the RFN-mediated regulation of riboflavin biosynthesis gene at the transcriptional level
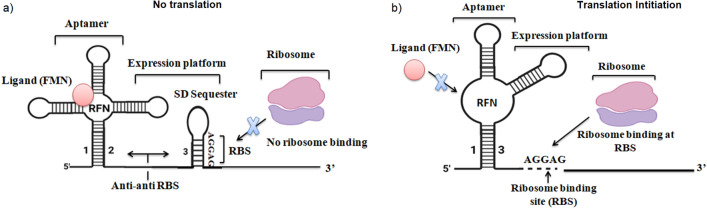
On the other hand, a fragment of the helix (mostly the left half) is effective to pair within an oligonucleotide of the aptamer domain (mostly the right half of the P1 helix) hence configuring anti-sequestering helix (Findeiß et al. 2017). This fundamental structural organization suggests that these riboswitches can regulate the initiation of translation efficiently by occluding Ribosomal Binding Site (RBS) under some specific conditions (Fig. 5a and b) (Chatterjee et al. 2021). Predominantly in Gram-negative bacteria, downstream of the RFN element, the RNA hairpins are used to sequester the RBS (SD sequence) (Yakhnin et al. 2006).
Fig. 5.
a and b The predicted attenuation mechanism of the RFN mediated regulation of riboflavin biosynthesis gene at translation level
Mostly, highly evolutionary conserved sequence GCCCTGA, are reported which are overlaps with the prospective sequester hairpin and showed complementary sequence with the base stem of the RFN element (Wilt et al. 2020). RNAs always used these two complementary sequences to form secondary structures, anti-sequester, and these structures are highly stable than the RFN element (Steinert et al. 2017). Probably, the proposed mechanism of regulation of initiation of translation of the riboflavin biosynthesis genes (rib operon) is closely similar to the mechanism as discussed above (termination–anti-termination), but in this mechanism, SD-sequester (Shine-Dalgarno sequence) is involved (Chandra et al. 2017) rather than terminator or terminator hairpin loops. Under repressive conditions, the RFN prevents the formation of the anti-sequester, and the anti-sequester as well as SD-sequester inhibit the initiation of translation. In the de-repressing circumstances, RFN is replaced by an anti-sequester, as a result, SD-box is released that permits translation to begin (Chatterjee et al. 2021). All riboswitches are selectively bound by their corresponding ligand and control gene expression if any removal and addition of a single functional group typically cause a less binding affinity (Jones and Ferré-D’Amaré 2017).
Riboswitch-mediated genetic control at the transcriptional as well as translational levels improves the metabolic stability and survival strategy in bacteria and encourages us to develop biotechnologically favorable strains using genetic engineering techniques. The mutated strain can be used for the synthesis of a particular metabolite such as riboflavin (vitB2) and cobalamin (vitB12) at the commercial level via biotechnology (Sherlock et al. 2018). Hence, some metabolic effects of different riboswitch-mediated regulations are discussed as follows:
FMN riboswitch
The FMN riboswitch is more complex and more widespread in bacteria and binds with FMN. In this riboswitch, the RFN element controls the riboflavin (vitB2) biosynthesis and transport genes. When RNA is incubated with FMN, it undergoes structural change due to the binding of FMN without requiring any proteins (Panchal and Brenk 2021). Part of this increase in structural complexity might be required for the RNA to make productive interactions with the phosphate moiety of FMN. The similar chemical riboflavin, which differs from FMN by the absence of a single phosphate group, binds almost 100-fold more tightly than the riboswitch found in the 5’ UTR region of the ribDEATH operon in Bacillus subtilis (Winkler and Breaker 2005).
Coenzyme B12 riboswitch
Numerous bacterial species contain 1–13 representatives’ coenzyme B12 or adenosylcobalamin-specific riboswitches only in a single genome. This riboswitch controls the expression of cobalamin synthesis protein-producing genes, but it may also regulate the expression of porphyrin and cobalt transport genes (Osman et al. 2021). In specific situations, this riboswitch controls the expression of proteins for succinate, glutamate and coenzyme B12 independent ribonucleotide reductase (Nahvi et al. 2002). The largest aptamer domains of all-natural aptamers discovered to date are often found in members of this class of riboswitches, indicating that the structural requirements are high for an RNA that is selectively recognizing this huge and chemically complex metabolite (Garst et al. 2011).
Lysine riboswitch
Another class of riboswitches that preferentially binds to L-lysine also seems to contain the k-turn motif. Again, this structural component is probably crucial for this RNA to fold extended stem − bulge elements into a more compact RNA structure, as does another frequent RNA motif known as loop E (Serganov and Patel 2009). Although the minimal lysine-binding aptamer from the Bacillus subtilis lycC mRNA has a low affinity (Kd = 1 µM), this low affinity may be due to the large size and extensive nucleotide conservation of lysine aptamers, which may be required to create a structure that excludes the numerous close analogous of lysine that is found in the majority of cells (Elskens et al. 2020). For instance, the aptamer distinguishes between the l and d forms of lysine and disqualifies substances like ornithine, homolysine and 5-hydroxylysine because they have a different side chain length or structure than l-lysine. The compound S-2 aminoethyl l-cysteine (AEC) is one l-lysine analog that the riboswitch binds to with a respectable affinity (~ 30 µM) (Wickiser et al. 2005). AEC is an antimetabolite that is poisonous to many bacteria and AEC-resistant strains of Bacillus subtilis possess mutations that impair the riboswitches ability to function and this mechanism makes this observation noteworthy (Blount et al. 2007). Although, the incorporation of AEC into proteins during translation (in place of lysine) might produce the antimicrobial effects observed in bacteria, the fact that AEC also activates riboswitch function suggests that at least some of the antimicrobial action of this compound could be attributed to the repression of genes involved in lysine biosynthesis (Machtel et al. 2016). Similarly, the other riboswitches could also be used as inventive drug targets to develop new classes of antibiotics.
Glycine riboswitch
In a wide range of bacteria, two remarkably comparable RNA structures were conserved upstream of the glycine catabolic and efflux genes (Khani et al. 2018). These domains are often located in the 5′ UTR of genes that encode the glycine cleavage machinery, which degrades glycine when there is an excess of this amino acid present (Serganov and Patel 2009). It was later discovered through biochemical studies utilizing creations from the bacteria Bacillus subtilis and Vibrio cholera that each conserved domain is associated with a single glycine molecule. Surprisingly, these aptamers reject chemical compounds with a similar structure, like alanine, serine and dipeptide glycylglycine (Torgerson et al. 2020). It is even more remarkable to see that once glycine binds to one aptamer domain, the second domain displays a cooperative increase in glycine affinity of at least 1000-fold (Butler et al. 2011). By variations in metabolite concentration that are much smaller than those seen with other riboswitches, this cooperative mechanism enables to control of gene expression across its whole range (Garst and Batey 2009). The microorganisms that use a cooperative riboswitch for glycine sensing may have a clear evolutionary advantage because this particular class is typically used for controlling glycine catabolism, in which over-expression would have detrimental effects (Mandal et al. 2004). With the help of a digital riboswitch, cells can maximally express glycine cleavage system proteins when there is a slight excess of glycine available and maximally suppress the expression of these proteins when glycine concentration is getting close to the critical limit for their critical use in protein synthesis (Elskens et al. 2020).
Techniques used to study riboswitch-mediated regulation
Riboswitches are dynamic RNA patterns, most frequently found in the 5′ UTR region of bacterial mRNAs that regulate the transcription and translation of genes by undergoing conformational changes upon binding of a ligand or metabolite (Wilt et al. 2020). The small size of a typical ligand or metabolite means that only a little amount of free energy is available from ligand binding to break through the frequently high energetic barrier of modifying RNA structure. Instead, most riboswitches seem to use the ligand as a structural “linchpin” to change the kinetic partitioning between different folds, using the directed and hierarchical folding of RNA (Wu and D’souza 2020). Since they avoid the ensemble averaging of bulk techniques, which obscures unsynchronized, transitory and/or multi-state kinetic behavior, the single-molecule (SM) microscopy technology is especially suited to explore such kinetically regulated RNA folding (Wu et al. 2012). In response to the binding of a metabolite or ligand that is small as compared to the riboswitch and/or especially the gene expression machinery, the riboswitch often undergo allosteric structural rearrangements (Sherwood et al. 2018). Because of this, the amount of binding energy released through the net gain of just a few kcal/mol of hydrogen bonds, ion–ion and/or stacking interaction might be insufficient to overcome the potential high (tens to hundreds of kcal/mol) free energy barriers to interconvert alternate RNA secondary structure (Suddala et al. 2018). Notably, the ligand frequently attaches as a structural “linchpin” to condense the aptamer and topologically close distal RNA segments. Because of the directional and hierarchical co-transcriptional folding of RNA, this may enable riboswitches to change the kinetic partitioning between various RNA folds to bring distal sequence fragments closer during transcription (Jones and Ferré-D’Amaré 2017). A branch point in the folding landscape where a little free energy bias can significantly affect the final secondary structure within an acceptable time may be exploited (i.e. seconds rather than days). SM approaches are frequently necessary for a quantitative description of the folding and unfolding of dynamic structural motifs of riboswitches (Zhang and Ferré-D’Amaré 2014). The key determinants of whether the downstream gene (s) are expressed, more specifically the kinetic variations of the riboswitch folding in the absence and presence of ligand relative to the kinetics of the cellular gene expression machinery. Rapid uptake of SM approaches for studying riboswitch processes is a result of their ability to specifically detect and quantify rare molecular events, transiently, visited states and different kinetic characteristics (Helmling et al. 2018).
In conclusion, the participation of non-coding RNAs and untranslated regions of mRNAs over gene control mechanisms seems to be more considerable than initially believed. Several mechanisms involved in the expression of small RNAs specifically recognize mRNAs and regulate the gene expression of that particular mRNAs. These non-coding RNAs or gene control factors could easily appear through the evolutionary mechanisms, provided that contingent base-pairing interactions with mRNAs influence cellular gene expression. These non-coding RNAs, and riboswitches form a binding pocket for particular metabolites and carry an essential domain that allosterically responds to metabolite binding. To perform these functions there is a need to attain specific structures by RNAs, which suggest that riboswitches contain highly evolutionarily conserved sequences. However, natural and artificial aptamers (engineered aptamers) are shows high affinity and specificity for a selected compound. In contrast, this cannot easily be precluding that ligands have not been changed over billions of years. In addition, the biochemical and structural studies of riboswitches help to understand how non-coding RNAs folded into a functional unit and allow us to make a clear picture of the structures and functions of non-coding RNAs and riboswitches.
Data availability statement
The data collected to write this review will be made available by the authors, without undue reservation, to any qualified researcher.
Author contributions
Conceptualization: VM and VK. Writing—original draft preparation: VK and VM. Writing—VK, VM, AR and JJA. Resources: VM and VK. Supervision: VM and JJA. All authors read and approved the manuscript.
Funding
The work was funded by National Institute of Food Technology Entrepreneurship and Management (NIFTEM), Sonipat, India.
Code availability
Not applicable.
Declarations
Conflict of interest
Dr. J J Ahire is employed at Unique Biotech Ltd., Hyderabad. Unique Biotech Limited had no role in the design/analysis/writing of the review. Other authors have no conflict of interest to declare.
Research involving human participants and/or animals
The paper does not contain any study on human participants or animals.
Informed consent
Not applicable.
Consent for publication
Not applicable.
Footnotes
Vijendra Mishra: Deceased on 6th March 2022.
References
- Abduljalil JM. Bacterial riboswitches and RNA thermometers: nature and contributions to pathogenesis. Non-Coding RNA Res. 2018;3:54–63. doi: 10.1016/j.ncrna.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson BW, Liu K, Wolak C, Dubiel K, She F, Satyshur KA, Keck JL, Wang JD. Evolution of (P)ppGpp-HPRT regulation through diversification of an allosteric oligomeric interaction. Elife. 2019;8:1–31. doi: 10.7554/eLife.47534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averianova LA, Balabanova LA, Son OM, Podvolotskaya AB, Tekutyeva LA. Production of vitamin B2 (riboflavin) by microorganisms: an overview. Front Bioeng Biotechnol. 2020 doi: 10.3389/fbioe.2020.570828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramaniyam T, Oh KI, Jin HS, Bin AH, Kim BS, Lee JH. Non-canonical helical structure of nucleic acids containing base-modified nucleotides. Int J Mol Sci. 2021 doi: 10.3390/ijms22179552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007 doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT. Structure and mechanism of purine-binding riboswitches. Q Rev Biophys. 2012;45:345–381. doi: 10.1017/S0033583512000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bervoets I, Charlier D. Diversity, versatility and complexity of bacterial gene regulation mechanisms: opportunities and drawbacks for applications in synthetic biology. FEMS Microbiol Rev. 2019;43:304–339. doi: 10.1093/femsre/fuz001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagdikar D, Grundy FJ, Henkin TM. Transcriptional and translational S-box riboswitches differ in ligand-binding properties. J Biol Chem. 2020;295:6849–6860. doi: 10.1074/jbc.RA120.012853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittrich S, Burley SK, Rose AS. Real-time structural motif searching in proteins using an inverted index strategy. PLoS Comput Biol. 2020;16:1–17. doi: 10.1371/journal.pcbi.1008502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nat Chem Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- Bou-Nader C, Zhang J. Structural insights into RNA dimerization: motifs, interfaces and functions. Molecules. 2020 doi: 10.3390/molecules25122881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler EB, Xiong Y, Wang J, Strobel SA. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol. 2011;18:293–298. doi: 10.1016/j.chembiol.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Heel AJ, Ahmed H, Mols M, Kuipers OP. Cell surface engineering of Bacillus subtilis improves production yields of heterologously expressed α-amylases. Microb Cell Fact. 2017;16:1–9. doi: 10.1186/s12934-017-0674-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capozzi V, Russo P. Lactic acid bacteria producing B-group vitamins: a great potential for functional cereals products. Appl Microbiol Biotechnol. 2012 doi: 10.1007/s00253-012-4440-2. [DOI] [PubMed] [Google Scholar]
- Chandra V, Hannan Z, Xu H, Mandal M. Single-molecule analysis reveals multi-state folding of a guanine riboswitch. Nat Chem Biol. 2017;13:194–201. doi: 10.1038/nchembio.2252. [DOI] [PubMed] [Google Scholar]
- Chatterjee S, Chauvier A, Dandpat SS, Artsimovitch I, Walter NG. A translational riboswitch coordinates nascent transcription-translation coupling. Proc Natl Acad Sci U S A. 2021 doi: 10.1073/pnas.2023426118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chełkowska-Pauszek A, Kosiński JG, Marciniak K, Wysocka M, Bakowska-Żywicka K, Żywicki M. The role of rna secondary structure in regulation of gene expression in bacteria. Int J Mol Sci. 2021 doi: 10.3390/ijms22157845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Morita T, Gottesman S. Regulation of transcription termination of small RNAs and by small RNAs: molecular mechanisms and biological functions. Front Cell Infect Microbiol. 2019;9:1–9. doi: 10.3389/fcimb.2019.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, White EN, Brandt NL, Yu AM, Chen AA, Lucks JB. Cotranscriptional RNA strand exchange underlies the gene regulation mechanism in a purine-sensing transcriptional riboswitch. bioRxiv. 2021 doi: 10.1101/2021.10.25.465737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillón I, Marcia M. The molecular structure of long non-coding RNAs: emerging patterns and functional implications. Crit Rev Biochem Mol Biol. 2020;55:662–690. doi: 10.1080/10409238.2020.1828259. [DOI] [PubMed] [Google Scholar]
- Di Salvo M, Puccio S, Peano C, Lacour S, Alifano P. RhoTermPredict: An algorithm for predicting Rho-dependent transcription terminators based on Escherichia coli, Bacillus subtilis and Salmonella enterica databases. BMC Bioinformatics. 2019;20:1–11. doi: 10.1186/s12859-019-2704-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogalis LK, Batey RT. Requirements for efficient ligand-gated cotranscriptional switching in designed variants of the B. subtilis pbuE adenine-responsive riboswitch in E. coli. PLoS ONE. 2020;15:1–26. doi: 10.1371/journal.pone.0243155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elskens JP, Elskens JM, Madder A. Chemical modification of aptamers for increased binding affinity in diagnostic applications: current status and future prospects. Int J Mol Sci. 2020;21:1–31. doi: 10.3390/ijms21124522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espah Borujeni A, Mishler DM, Wang J, Huso W, Salis HM. Automated physics-based design of synthetic riboswitches from diverse RNA aptamers. Nucleic Acids Res. 2016;44:1–13. doi: 10.1093/nar/gkv1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findeiß S, Etzel M, Will S, Mörl M, Stadler PF. Design of artificial riboswitches as biosensors. Sensors (switzerland) 2017;17:1–28. doi: 10.3390/s17091990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes Flores A, Sepúlveda-Cisternas I, Vásquez Solis De Ovando JI, Torres A, García-Angulo VA. Contribution of riboflavin supply pathways to Vibrio cholerae in different environments. Gut Pathog. 2017;9:1–9. doi: 10.1186/s13099-017-0214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser LR, Kelly ML, Herschlag D, Al-hashimi HM, Chemistry SC, Chemistry SC, Chemistry SC (2020) HHS Public Access. 20:474–489. 10.1038/s41580-019-0136-0
- García-Angulo VA. Overlapping riboflavin supply pathways in bacteria. Crit Rev Microbiol. 2017;43:196–209. doi: 10.1080/1040841X.2016.1192578. [DOI] [PubMed] [Google Scholar]
- Garst AD, Batey RT. A switch in time: detailing the life of a riboswitch. Biochim Biophys Acta Gene Regul Mech. 2009;1789:584–591. doi: 10.1016/j.bbagrm.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Héroux A, Rambo RP, Batey RT. Crystal structure of the lysine riboswitch regulatory mRNA element. J Biol Chem. 2008;283:22347–22351. doi: 10.1074/jbc.C800120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Edwards AL, Batey RT. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol. 2011;3:1–13. doi: 10.1101/cshperspect.a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Marchisio MA. Aptamers, riboswitches and ribozymes in S. cerevisiae synthetic biology. Life. 2021 doi: 10.3390/life11030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Wang Y, Wang Z, Zhang W. Co-transcriptional folding and regulation mechanisms of riboswitches. Molecules. 2017;22:1–14. doi: 10.3390/molecules22071169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy FJ, Lehman SC, Henkin TM. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci U S A. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Pal D. Clusters of hairpins induce intrinsic transcription termination in bacteria. Sci Rep. 2021;11:1–18. doi: 10.1038/s41598-021-95435-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle V, Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol. 2018;19:621–637. doi: 10.1038/s41580-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann U, Roske Y. Symmetry in nucleic-acid double helices. Symmetry (basel) 2020 doi: 10.3390/SYM12050737. [DOI] [Google Scholar]
- Helmann JD. Where to begin? Sigma factors and the selectivity of transcription initiation in bacteria. Mol Microbiol. 2019;112:335–347. doi: 10.1111/mmi.14309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmling C, Klötzner DP, Sochor F, Mooney RA, Wacker A, Landick R, Fürtig B, Heckel A, Schwalbe H. Life times of metastable states guide regulatory signaling in transcriptional riboswitches. Nat Commun. 2018 doi: 10.1038/s41467-018-03375-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix DK, Brenner SE, Holbrook SR. RNA structural motifs: building blocks of a modular biomolecule. Q Rev Biophys. 2005;38:221–243. doi: 10.1017/S0033583506004215. [DOI] [PubMed] [Google Scholar]
- Henkin TM, Glass BL, Grundy FJ. Analysis of the Bacillus subtilis tyrS gene: conservation of a regulatory sequence in multiple tRNA synthetase genes. J Bacteriol. 1992;174:1299–1306. doi: 10.1128/jb.174.4.1299-1306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;80(287):820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- Hoffmann M, Braig A, Fernandez Cano Luna DS, Rief K, Becker P, Treinen C, Klausmann P, Morabbi Heravi K, Henkel M, Lilge L, Hausmann R. Evaluation of an oxygen-dependent self-inducible surfactin synthesis in B. subtilis by substitution of native promoter PsrfA by anaerobically active PnarG and PnasD. AMB Express. 2021 doi: 10.1186/s13568-021-01218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook SR. Structural principles from large RNAs. Annu Rev Biophys. 2008;37:445–464. doi: 10.1146/annurev.biophys.36.040306.132755. [DOI] [PubMed] [Google Scholar]
- Hua B, Jones CP, Mitra J, Murray PJ, Rosenthal R, Ferré-D’Amaré AR, Ha T. Real-time monitoring of single ZTP riboswitches reveals a complex and kinetically controlled decision landscape. Nat Commun. 2020;11:1–11. doi: 10.1038/s41467-020-18283-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst T, Xu X, Zhao P, Chen SJ. Quantitative understanding of SHAPE mechanism from RNA structure and dynamics analysis. J Phys Chem B. 2018;122:4771–4783. doi: 10.1021/acs.jpcb.8b00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husser C, Dentz N, Ryckelynck M. Structure-switching RNAs: from gene expression regulation to small molecule detection. Small Struct. 2021;2:2000132. doi: 10.1002/sstr.202000132. [DOI] [Google Scholar]
- Irastortza-Olaziregi M, Amster-Choder O. Coupled transcription-translation in prokaryotes: an old couple with new surprises. Front Microbiol. 2021 doi: 10.3389/fmicb.2020.624830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irla M, Hakvåg S, Brautaset T. Developing a riboswitch-mediated regulatory system for metabolic flux control in thermophilic Bacillus methanolicus. Int J Mol Sci. 2021 doi: 10.3390/ijms22094686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CP, Ferré-D’Amaré AR. Long-range interactions in riboswitch control of gene expression. Annu Rev Biophys. 2017;46:455–481. doi: 10.1146/annurev-biophys-070816-034042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khani A, Popp N, Kreikemeyer B, Patenge N. A glycine riboswitch in Streptococcus pyogenes controls expression of a sodium: alanine symporter family protein gene. Front Microbiol. 2018;9:1–10. doi: 10.3389/fmicb.2018.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kil YV, Mironovi VN, Gorishin IY, Kreneva RA, Perumov DA. Riboflavin operon of Bacillus subtilis: unusual symmetric arrangement of the regulatory region. MGG Mol Gen Genet. 1992;233:483–486. doi: 10.1007/BF00265448. [DOI] [PubMed] [Google Scholar]
- Kochhar S, Paulus H. Lysine-induced premature transcription termination in the lysC operon of Bacillus subtilis. Microbiol. 1996;142:1635–1639. doi: 10.1099/13500872-142-7-1635. [DOI] [PubMed] [Google Scholar]
- Kovach IM. Proton bridging in catalysis by and inhibition of serine proteases of the blood cascade system. Life. 2021 doi: 10.3390/life11050396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lah J, Seručnik M, Vesnaver G. Influence of a hairpin loop on the thermodynamic stability of a DNA oligomer. J Nucleic Acids. 2011 doi: 10.4061/2011/513910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara AR, Jaén KE, Sigala JC, Mühlmann M, Regestein L, Büchs J. Characterization of endogenous and reduced promoters for oxygen-limited processes using Escherichia coli. ACS Synth Biol. 2017;6:344–356. doi: 10.1021/acssynbio.6b00233. [DOI] [PubMed] [Google Scholar]
- Liberman JA, Suddala KC, Aytenfisu A, Chan D, Belashov IA, Salim M, Mathews DH, Spitale RC, Walter NG, Wedekind JE. Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics. Proc Natl Acad Sci U S A. 2015;112:E3485–E3494. doi: 10.1073/pnas.1503955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Chao YF, Weng SF. Riboflavin synthesis genes ribE, ribB, ribH, ribA reside in the lux operon of Photobacterium leiognathi. Biochem Biophys Res Commun. 2001;284:587–595. doi: 10.1006/bbrc.2001.5013. [DOI] [PubMed] [Google Scholar]
- Lins MRCR, Correa GG, Amorim LAS, Franco RAL, Ribeiro NV, de Jesus VN, Pedrolli DB. Characterization of five purine riboswitches in cellular and cell-free expression systems. bioRxiv. 2021 doi: 10.1101/2021.04.14.439898. [DOI] [PubMed] [Google Scholar]
- Machtel P, Bąkowska-Żywicka K, Żywicki M. Emerging applications of riboswitches—from antibacterial targets to molecular tools. J Appl Genet. 2016;57:531–541. doi: 10.1007/s13353-016-0341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machtel P, Wasilewska-burczyk A, Zacharjasz J, Kamilla BŻ (2021) PTT-quant—a new method for direct identification and absolute quantification of premature transcription termination events, following the example of bacterial riboswitches [DOI] [PMC free article] [PubMed]
- Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/S0092-8674(03)00391-X. [DOI] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;80(306):275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- Matyjasik MM, Batey RT. Structural basis for 2′-deoxyguanosine recognition by the 2′-dG-II class of riboswitches. Nucleic Acids Res. 2019;47:10931–10941. doi: 10.1093/nar/gkz839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR. Riboswitch diversity and distribution. RNA. 2017;23:995–1011. doi: 10.1261/rna.061234.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel BA, Grundy FJ, Henkin TM. A tertiary structural element in S box leader RNAs is required for S-adenosylmethionine-directed transcription termination. Mol Microbiol. 2005;57:1008–1021. doi: 10.1111/j.1365-2958.2005.04740.x. [DOI] [PubMed] [Google Scholar]
- Mehdizadeh Aghdam E, Sinn M, Tarhriz V, Barzegar A, Hartig JS, Hejazi MS. TPP riboswitch characterization in Alishewanella tabrizica and Alishewanella aestuarii and comparison with other TPP riboswitches. Microbiol Res. 2017;195:71–80. doi: 10.1016/j.micres.2016.11.003. [DOI] [PubMed] [Google Scholar]
- Miao Z, Westhof E. RNA Structure: advances and assessment of 3D structure prediction. Annu Rev Biophys. 2017;46:483–503. doi: 10.1146/annurev-biophys-070816-034125. [DOI] [PubMed] [Google Scholar]
- Micura R, Höbartner C. Fundamental studies of functional nucleic acids: aptamers, riboswitches, ribozymes and DNAzymes. Chem Soc Rev. 2020;49:7331–7353. doi: 10.1039/d0cs00617c. [DOI] [PubMed] [Google Scholar]
- Mironov VN, Kraev AS, Chikindas ML, Chernov BK, Stepanov AI, Skryabin KG (1994) DIG’-G. 201–208 [DOI] [PubMed]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem Biol. 2002;9:1043–1049. doi: 10.1016/S1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Noeske J, Schwalbe H, Wöhnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2007;35:5262–5273. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouwen N, Arrighi JF, Gully D, Giraud E. RibBX of bradyrhizobium ORS285 plays an important role in intracellular persistence in various aeschynomene host plants. Mol Plant-Microbe Interact. 2021;34:88–99. doi: 10.1094/MPMI-07-20-0209-R. [DOI] [PubMed] [Google Scholar]
- Novoseltseva AA, Zavyalova EG, Golovin AV, Kopylov AM. An insight into aptamer—protein complexes. Aptamers. 2018;2:55–63. [Google Scholar]
- Oliveira GP, dos Andrade ACSP, Rodrigues RAL, Arantes TS, Boratto PVM, Silva LKDS, Dornas FP, de Trindade GS, Drumond BP, La Scola B, Kroon EG, Abrahão JS. Promoter motifs in NCLDVs: an evolutionary perspective. Viruses. 2017;9:1–20. doi: 10.3390/v9010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr MW, Mao Y, Storz G, Qian SB. Alternative ORFs and small ORFs: shedding light on the dark proteome. Nucleic Acids Res. 2021;48:1029–1042. doi: 10.1093/NAR/GKZ734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman D, Cooke A, Young TR, Deery E, Robinson NJ, Warren MJ. The requirement for cobalt in vitamin B12: a paradigm for protein metalation. Biochim Biophys Acta - Mol Cell Res. 2021;1868:118896. doi: 10.1016/j.bbamcr.2020.118896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page K, Shaffer J, Lin S, Zhang M, Liu JM. Engineering riboswitches in vivo using dual genetic selection and fluorescence-activated cell sorting. ACS Synth Biol. 2018;7:2000–2006. doi: 10.1021/acssynbio.8b00099. [DOI] [PubMed] [Google Scholar]
- Panchal V, Brenk R. Riboswitches as drug targets for antibiotics. Antibiotics. 2021;10:1–22. doi: 10.3390/antibiotics10010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrolli D, Langer S, Hobl B, Schwarz J, Hashimoto M, Mack M. The ribB FMN riboswitch from Escherichia coli operates at the transcriptional and translational level and regulates riboflavin biosynthesis. FEBS J. 2015;282:3230–3242. doi: 10.1111/febs.13226. [DOI] [PubMed] [Google Scholar]
- Pedrolli DB, Kühm C, Sévin DC, Vockenhuber MP, Sauer U, Suess B, Mack M. A dual control mechanism synchronizes riboflavin and sulphur metabolism in Bacillus subtilis. Proc Natl Acad Sci U S A. 2015;112:14054–14059. doi: 10.1073/pnas.1515024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polaski JT, Holmstrom ED, Nesbitt DJ, Batey RT. Mechanistic insights into cofactor-dependent coupling of RNA folding and mRNA transcription/translation by a cobalamin riboswitch. Cell Rep. 2016;15:1100–1110. doi: 10.1016/j.celrep.2016.03.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Chauvier A, Walter NG. Kinetics coming into focus: single-molecule microscopy of riboswitch dynamics. RNA Biol. 2019;16:1077–1085. doi: 10.1080/15476286.2018.1536594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberi F, Kamali M, Najafi A, Yazdanparast A, Moghaddam MM. Natural antisense RNAs as mRNA regulatory elements in bacteria: a review on function and applications. Cell Mol Biol Lett. 2016;21:1–17. doi: 10.1186/s11658-016-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder KT, Daldrop P, Lilley DMJ. RNA tertiary interactions in a riboswitch stabilize the structure of a kink turn. Structure. 2011;19:1233–1240. doi: 10.1016/j.str.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scull CE, Dandpat SS, Romero RA, Walter NG. Transcriptional riboswitches integrate timescales for bacterial gene expression control. Front Mol Biosci. 2021;7:1–10. doi: 10.3389/fmolb.2020.607158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Amino acid recognition and gene regulation by riboswitches. Biochim Biophys Acta Gene Regul Mech. 2009;1789:592–611. doi: 10.1016/j.bbagrm.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–237. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw GC, Fulco AJ. Barbiturate-mediated regulation of expression of the cytochrome P450(BM-3) gene of Bacillus megaterium by Bm3R1 protein. J Biol Chem. 1992;267:5515–5526. doi: 10.1016/s0021-9258(18)42797-4. [DOI] [PubMed] [Google Scholar]
- Sherlock ME, Sudarsan N, Stav S, Breaker RR. Tandem riboswitches form a natural Boolean logic gate to control purine metabolism in bacteria. Elife. 2018;7:1–17. doi: 10.7554/eLife.33908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood AV, Frandsen JK, Grundy FJ, Henkin TM. New tRNA contacts facilitate ligand binding in a Mycobacterium smegmatis T box riboswitch. Proc Natl Acad Sci U S A. 2018;115:3894–3899. doi: 10.1073/pnas.1721254115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Yamazaki Y, Tanaka K, Ishihama A. The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli. PLoS ONE. 2014 doi: 10.1371/journal.pone.0090447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinumvayo JP, Zhao C, Tuyishime P. Recent advances and future trends of riboswitches: attractive regulatory tools. World J Microbiol Biotechnol. 2018 doi: 10.1007/s11274-018-2554-0. [DOI] [PubMed] [Google Scholar]
- Sklyarova SA, Kreneva RA, Perumov DA, Mironov AS. The characterization of internal promoters in the Bacillus subtilis riboflavin biosynthesis operon. Russ J Genet. 2012;48:967–974. doi: 10.1134/S1022795412100109. [DOI] [PubMed] [Google Scholar]
- Steinert H, Sochor F, Wacker A, Buck J, Helmling C, Hiller F, Keyhani S, Noeske J, Grimm S, Rudolph MM, Keller H, Mooney RA, Landick R, Suess B, Fürtig B, Wöhnert J, Schwalbe H. Pausing guides RNA folding to populate transiently stable RNA structures for riboswitch-based transcription regulation. Elife. 2017;6:1–18. doi: 10.7554/eLife.21297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel EJ, Cheng L, Berman KE, Carlson PD, Julius B, Engineering B (2020) Riboswitch transcription control. 15:1067–1076. 10.1038/s41589-019-0382-7
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic Di-GMP. Science. 2008;80(321):411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suddala KC, Cabello-Villegas J, Michnicka M, Marshall C, Nikonowicz EP, Walter NG. Hierarchical mechanism of amino acid sensing by the T-box riboswitch. Nat Commun. 2018;9:1–14. doi: 10.1038/s41467-018-04305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudzinová P, Kambová M, Ramaniuk O, Benda M, Šanderová H, Krásný L. Effects of DNA topology on transcription from RRNA promoters in Bacillus subtilis. Microorganisms. 2021;9:1–17. doi: 10.3390/microorganisms9010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur K, Tomar SK, Wei ZJ. Comparative mRNA expression profiles of riboflavin biosynthesis genes in lactobacilli isolated from human feces and fermented bamboo shoots. Front Microbiol. 2017;8:427. doi: 10.3389/fmicb.2017.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tickner ZJ, Farzan M. Riboswitches for controlled expression of therapeutic transgenes delivered by adeno-associated viral vectors. Pharmaceuticals. 2021;14:1–29. doi: 10.3390/ph14060554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torgerson CD, Hiller DA, Strobel SA. The asymmetry and cooperativity of tandem glycine riboswitch aptamers. RNA. 2020;26:564–580. doi: 10.1261/rna.073577.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogl C, Grill S, Schilling O, Stülke J, Mack M, Stolz J. Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum. J Bacteriol. 2007;189:7367–7375. doi: 10.1128/JB.00590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Monford Paul Abishek N, Jeon HJ, Lee Y, He J, Adhya S, Lim HM. Processing generates 3′ ends of RNA masking transcription termination events in prokaryotes. Proc Natl Acad Sci U S A. 2019;116:4440–4445. doi: 10.1073/pnas.1813181116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Wang Z, Pan N, Huang J, Wan C. Improved identification of small open reading frames encoded peptides by top–down proteomic approaches and de novo sequencing. Int J Mol Sci. 2021 doi: 10.3390/ijms22115476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Nelson JW, Lönse CE, Sherlock ME, Breaker RR. Bioinformatic analysis of riboswitch structures uncovers variant classes with altered ligand specificity. Proc Natl Acad Sci U S A. 2017;114:E2077–E2085. doi: 10.1073/pnas.1619581114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Wilt HM, Yu P, Tan K, Wang YX, Stagno JR. FMN riboswitch aptamer symmetry facilitates conformational switching through mutually exclusive coaxial stacking configurations. J Struct Biol X. 2020;4:100035. doi: 10.1016/j.yjsbx.2020.100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Wu MTP, D’souza V. Alternate RNA structures. Cold Spring Harb Perspect Biol. 2020;12:1–17. doi: 10.1101/cshperspect.a032425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Chai D, Fraser ME, Zimmerly S. Structural variation and uniformity among tetraloop-receptor interactions and other loop-helix interactions in RNA crystal structures. PLoS ONE. 2012 doi: 10.1371/journal.pone.0049225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin H, Yakhnin AV, Babitzke P. The trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis regulates translation initiation of ycbK, a gene encoding a putative efflux protein, by blocking ribosome binding. Mol Microbiol. 2006;61:1252–1266. doi: 10.1111/j.1365-2958.2006.05278.x. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ferré-D’Amaré AR. Direct evaluation of tRNA aminoacylation status by the T-Box riboswitch using tRNA−mRNA stacking and steric readout. Mol Cell. 2014;55:148–155. doi: 10.1016/j.molcel.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-R, Ge Y-Y, Liu P-H, Wu D-T, Liu H-Y, Li H-B, Corke H, Gan R-Y. Biotechnological strategies of riboflavin biosynthesis in microbes. Engineering. 2021 doi: 10.1016/j.eng.2021.03.018. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data collected to write this review will be made available by the authors, without undue reservation, to any qualified researcher.
Not applicable.