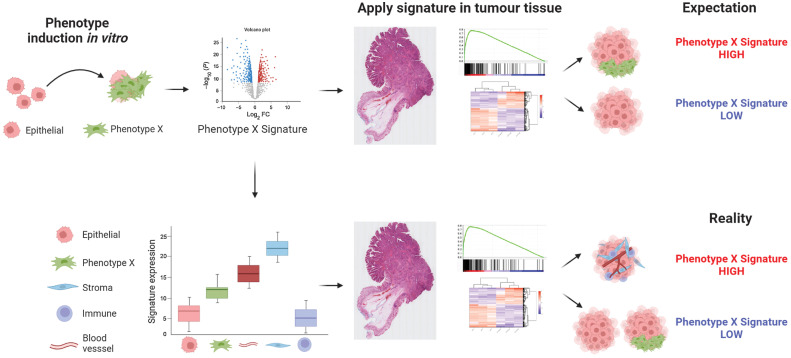
Figure 7.
Summary diagram. Precise gene expression signatures have been developed to accurately reflect phenotypic changes in epithelial-based models, when assessed under tightly controlled in vitro modeling conditions. When these signatures are used to stratify bulk tumor data, there is an expectation that the same signatures can be used to stratify tumors based on the same distinct phenotypes (top). However, if the genes that make up these signatures are expressed at relatively higher levels in nonepithelial lineages, the signatures can become confounded by even small variations in stromal components, stratifying tumors based on stromal content rather than the phenotype they were developed to represent (bottom).