Figure 3.
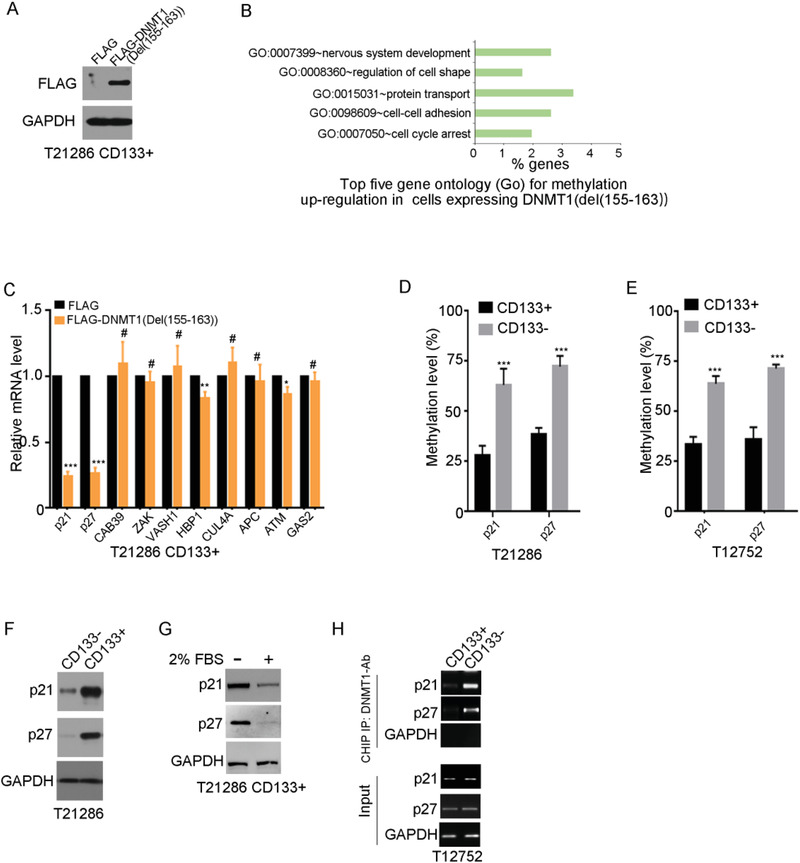
The CD133–DNMT1 interaction upregulates p21 and p27. A) Western blot analysis of FLAG‐DNMT1 in CD133+ cells expressing FLAG or FLAG‐DNMT1(Del(155–163)). GAPDH is used as a loading control. B) By Infinium MethylationEPIC BeadChip arrays, the methylation of 680 annotated genes is increased in CD133+ cells expressing DNMT1 (Del(155–163)) compared to control cells (p < 0.001, Δβ ≥ 0.15). Gene ontology results (top five, according to p value) for 680 genes in which methylation is upregulated in CD133+ cells expressing FLAG‐DNMT1(Del(155–163)) are shown. C) qRT‐PCR quantification of the indicated gene mRNA levels in T21286 CD133+ cells expressing FLAG or FLAG‐DNMT1(Del(155–163)). Data is shown as mean ± SD from three independent experiments; ***p < 0.001, **p < 0.01, *p < 0.05, #p > 0.05, Student's t‐test. D‐E) The methylation rate of p21 and p27 promoters in CD133+ cells and matched CD133‐ cells from T21286 (D) and T12752 (E) are analyzed by bisulfite sequencing. Methylation levels are determined by the ratio of converted C nucleotides to total C nucleotides following bisulfite treatment under CpG island. Results are expressed as mean ± SD from three independent experiments; ***p < 0.001, Student's t‐test. F) Western blot analysis of p21 and p27 expression in T21286 CD133+ cells and CD133‐ cells. GAPDH is used as a loading control. G) Western blot analysis of p21 and p27 expression in T21286 CD133+ cells treated with 2% FBS for 7 days. GAPDH is used as a loading control. H) Chromatin immunoprecipitation (ChIP) assay is performed in CD133+ cells and CD133‐ cells using a DNMT1 specific antibody, followed by PCR amplification of p21 and p27 promoter regions between +250 to position −100. Chromatin (defined as input) and GAPDH products immunoprecipitated by DNMT1 Ab are used as positive and negative control.
